| Sequence ID | X_DroMel_CAF1 |
|---|---|
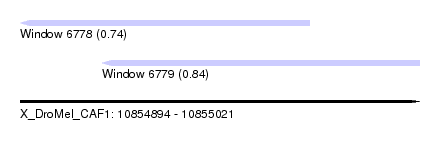
| Location | 10,854,894 – 10,855,021 |
| Length | 127 |
| Max. P | 0.842960 |

| Location | 10,854,894 – 10,854,986 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 72.08 |
| Mean single sequence MFE | -17.09 |
| Consensus MFE | -2.39 |
| Energy contribution | -2.83 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.14 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
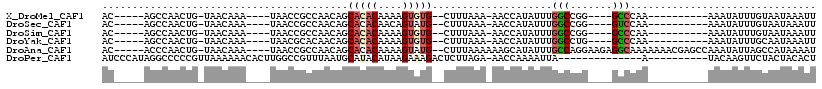
>X_DroMel_CAF1 10854894 92 - 22224390 AC-----AGCCAACUG-UAACAAA----UAACCGCCAACAGCACACAAAAGUGUG--CUUUAAA-AACCAUAUUUGGCCGG----GCCCAA----------AAAUAUUUGUAAUAAAUU ((-----((....)))-).(((((----((...(((...(((((((....)))))--)).....-..(((....)))...)----))....----------...)))))))........ ( -21.40) >DroSec_CAF1 8757 92 - 1 AC-----AGCCAACUG-UAACAAA----UAACCGCCAACAGCACACAACAGUAUG--CUUUAAA-AACCAUAUUUGGCCGG----GUCCAA----------AAAUAUUUGUAAUAAAUU ((-----((....)))-).(((((----((((((((((.((((.((....)).))--)).....-........)))))..)----))....----------...)))))))........ ( -14.65) >DroSim_CAF1 8868 92 - 1 AC-----AGCCAACUG-UAACAAA----UAACCGCCAACAGCACACAAAAGUGUG--CUUUAAA-AACCAUAUUUGGCCGG----GCCCAA----------AAAUAUUUGUAAUAAAUU ((-----((....)))-).(((((----((...(((...(((((((....)))))--)).....-..(((....)))...)----))....----------...)))))))........ ( -21.40) >DroYak_CAF1 8904 92 - 1 AC-----AGCCAACUG-UAACAAA----UAACGCACAACAGCACACAAAAGUGUG--CUUUAAA-AACCAUAUUUGGCCUG----GCCCAA----------AAAUAUUUGCAAUAAAUU ((-----((....)))-)..((((----((.........(((((((....)))))--)).....-.......(((((....----..))))----------)..))))))......... ( -18.30) >DroAna_CAF1 3752 107 - 1 AC-----ACCCAACUG-UAACAAA----UAACCGCCAACAGCACACAAAAGUAUG--CUUUAAAAAAGCAUAUUUGCCAGGAAGAGGCAAAAAAACGAGCCAAAUAUUAGCCAUAAAAU ..-----......(((-.......----.....((.....))......(((((((--(((.....))))))))))..))).....(((..........))).................. ( -15.70) >DroPer_CAF1 27622 94 - 1 AUCCCAUAGGCCCCCGUUAAAAAACACUUGGCCGUUUAAUGCAUACAUAAGAAAGACUCUUAGA-AACCAAAAUUA--------------A----------UACAAGUUCUACUACACU ........((((...(((....)))....))))(((((((.......(((((.....)))))..-.......))))--------------)----------.))............... ( -11.09) >consensus AC_____AGCCAACUG_UAACAAA____UAACCGCCAACAGCACACAAAAGUAUG__CUUUAAA_AACCAUAUUUGGCCGG____GCCCAA__________AAAUAUUUGUAAUAAAUU .........................................(((((....)))))....................(((.......)))............................... ( -2.39 = -2.83 + 0.45)

| Location | 10,854,920 – 10,855,021 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.74 |
| Mean single sequence MFE | -21.21 |
| Consensus MFE | -3.38 |
| Energy contribution | -4.85 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.16 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10854920 101 - 22224390 CACUAA-AAAGCCAGGCGGCCCA----GGCCAAUCAAAGAAC-----AGCCAACUG-UAACAAA----UAACCGCCAACAGCACACAAAAGUGUG--CUUUAAA-AACCAUAUUUGGCC ......-...((((((((((...----.))).........((-----((....)))-)......----.....)))...(((((((....)))))--)).....-.........)))). ( -30.00) >DroSec_CAF1 8783 98 - 1 CACUGA-AAAGCCAGG---CCCA----GGCCAAUCAAAGAAC-----AGCCAACUG-UAACAAA----UAACCGCCAACAGCACACAACAGUAUG--CUUUAAA-AACCAUAUUUGGCC ......-...((((((---(...----.))).........((-----((....)))-)......----...........((((.((....)).))--)).....-.........)))). ( -17.50) >DroSim_CAF1 8894 98 - 1 CACUGA-AAAGCCAGG---CCCA----GGCCAAUCAAAGUAC-----AGCCAACUG-UAACAAA----UAACCGCCAACAGCACACAAAAGUGUG--CUUUAAA-AACCAUAUUUGGCC ..((..-..))...((---((..----(((.........(((-----((....)))-)).....----.....)))...(((((((....)))))--)).....-..........)))) ( -25.11) >DroYak_CAF1 8930 99 - 1 CACUGAAAAAGCCAGG---CCCA----GGCCAAUCAAAGUAC-----AGCCAACUG-UAACAAA----UAACGCACAACAGCACACAAAAGUGUG--CUUUAAA-AACCAUAUUUGGCC ..........((((((---(...----.)))........(((-----((....)))-)).....----...........(((((((....)))))--)).....-.........)))). ( -24.80) >DroAna_CAF1 3792 92 - 1 ---UUA-AA----UGG---CCCA----AGCCAAUCAAAAAAC-----ACCCAACUG-UAACAAA----UAACCGCCAACAGCACACAAAAGUAUG--CUUUAAAAAAGCAUAUUUGCCA ---...-..----(((---(...----.))))..........-----......(((-(......----.........)))).......(((((((--(((.....)))))))))).... ( -14.66) >DroPer_CAF1 27641 104 - 1 ---UUA-AG----AGG---CCCAGACAGUCCAUUCAUUGAAUCCCAUAGGCCCCCGUUAAAAAACACUUGGCCGUUUAAUGCAUACAUAAGAAAGACUCUUAGA-AACCAAAAUUA--- ---(((-((----((.---............((.((((((((......((((...(((....)))....)))))))))))).))............))))))).-...........--- ( -15.21) >consensus CACUGA_AAAGCCAGG___CCCA____GGCCAAUCAAAGAAC_____AGCCAACUG_UAACAAA____UAACCGCCAACAGCACACAAAAGUAUG__CUUUAAA_AACCAUAUUUGGCC ..........(((((............(((..................)))..............................(((((....)))))..................))))). ( -3.38 = -4.85 + 1.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:58 2006