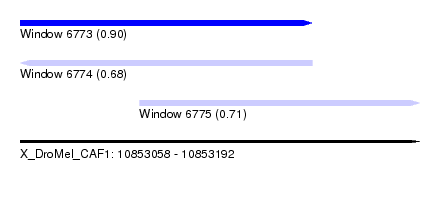
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,853,058 – 10,853,192 |
| Length | 134 |
| Max. P | 0.901961 |

| Location | 10,853,058 – 10,853,156 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -23.91 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10853058 98 + 22224390 CCCCAAUGUGUUAUGCCAAAAGCUAAAAACUUAUGCUUUUGCCAGUUUAUUGUCUUGUUUUGCUU-UGUCUGGUGCGACAGGCAGGCAGCAAAACAAAG ...(((((.....((.(((((((...........))))))).))...)))))..(((((((((((-((((((......)))))))..)))))))))).. ( -30.50) >DroSec_CAF1 6950 97 + 1 CUCCAAUGUGUUAUGCCAAAAGCUAAAAACUUAUGCUUUUGCCAGUUUAUUGUCUUGUUUUGCUU-UGUCU-GUGCGACAGGCAGGCAGCAAAACAAAG ...(((((.....((.(((((((...........))))))).))...)))))..(((((((((((-(((((-(.....)))))))..)))))))))).. ( -29.80) >DroSim_CAF1 7058 97 + 1 CCCCAAUGUGUUAUGCCAAAAGCUAAAAACUUAUGCUUUUGCCAGUUUAUUGUCUUGUUUUGCUU-UGUCU-GUGCGACAGGCAGGCGGCAAAACAAAG ...(((((.....((.(((((((...........))))))).))...)))))..(((((((((((-(((((-(.....)))))))..)))))))))).. ( -29.80) >DroYak_CAF1 7075 94 + 1 CCCCAAUGUGUUAUGCCAAAAGCUAAAAACUUAUGCUUUUGCCAGUUUAUUGUCUUGUUUUGCUUUUGUCU-GUGCG----GCAGGCAGCCAAACAAAG ...(((((.....((.(((((((...........))))))).))...)))))..((((((.(((...((((-(....----.)))))))).)))))).. ( -23.40) >consensus CCCCAAUGUGUUAUGCCAAAAGCUAAAAACUUAUGCUUUUGCCAGUUUAUUGUCUUGUUUUGCUU_UGUCU_GUGCGACAGGCAGGCAGCAAAACAAAG ...(((((.....((.(((((((...........))))))).))...)))))..((((((((((..(((((........)))))...)))))))))).. (-23.91 = -24.48 + 0.56)



| Location | 10,853,058 – 10,853,156 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.67 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10853058 98 - 22224390 CUUUGUUUUGCUGCCUGCCUGUCGCACCAGACA-AAGCAAAACAAGACAAUAAACUGGCAAAAGCAUAAGUUUUUAGCUUUUGGCAUAACACAUUGGGG .(((((((((((...((.(((......))).))-.))))))))))).((((....((.(((((((...........))))))).))......))))... ( -25.40) >DroSec_CAF1 6950 97 - 1 CUUUGUUUUGCUGCCUGCCUGUCGCAC-AGACA-AAGCAAAACAAGACAAUAAACUGGCAAAAGCAUAAGUUUUUAGCUUUUGGCAUAACACAUUGGAG .(((((((((((...((.(((.....)-)).))-.))))))))))).((((....((.(((((((...........))))))).))......))))... ( -26.10) >DroSim_CAF1 7058 97 - 1 CUUUGUUUUGCCGCCUGCCUGUCGCAC-AGACA-AAGCAAAACAAGACAAUAAACUGGCAAAAGCAUAAGUUUUUAGCUUUUGGCAUAACACAUUGGGG .((((((((((....((.(((.....)-)).))-..)))))))))).((((....((.(((((((...........))))))).))......))))... ( -23.30) >DroYak_CAF1 7075 94 - 1 CUUUGUUUGGCUGCCUGC----CGCAC-AGACAAAAGCAAAACAAGACAAUAAACUGGCAAAAGCAUAAGUUUUUAGCUUUUGGCAUAACACAUUGGGG .((((((((((.(....)----.)).)-)))))))............((((....((.(((((((...........))))))).))......))))... ( -20.20) >consensus CUUUGUUUUGCUGCCUGCCUGUCGCAC_AGACA_AAGCAAAACAAGACAAUAAACUGGCAAAAGCAUAAGUUUUUAGCUUUUGGCAUAACACAUUGGGG .(((((((((((.......((((......))))..))))))))))).((((....((.(((((((...........))))))).))......))))... (-21.42 = -22.67 + 1.25)


| Location | 10,853,098 – 10,853,192 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.21 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -18.11 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10853098 94 + 22224390 GCCAGUUUAUUGUCUUGUUUUGCUU-UGUCUGGUGCGACAGGCAGGCAGCAAAACAAAGUGGAAAACUCUUUUUCCAGCACCCAAAA----AAAUAUUC ((............(((((((((((-((((((......)))))))..))))))))))..(((((((....)))))))))........----........ ( -27.10) >DroSec_CAF1 6990 97 + 1 GCCAGUUUAUUGUCUUGUUUUGCUU-UGUCU-GUGCGACAGGCAGGCAGCAAAACAAAGUGGAAAACUCUUUUUCCAGCACCCAAAAAAAAAAAUAUUC ((............(((((((((((-(((((-(.....)))))))..))))))))))..(((((((....))))))))).................... ( -26.40) >DroSim_CAF1 7098 96 + 1 GCCAGUUUAUUGUCUUGUUUUGCUU-UGUCU-GUGCGACAGGCAGGCGGCAAAACAAAGUGGAAAACUCUUUUCCCAGCACCCAAAAA-AAAAAUAUUC ((............(((((((((((-(((((-(.....)))))))..)))))))))).(.(((((.....)))))).)).........-.......... ( -24.40) >DroYak_CAF1 7115 89 + 1 GCCAGUUUAUUGUCUUGUUUUGCUUUUGUCU-GUGCG----GCAGGCAGCCAAACAAAGUGGGAAACUCUUUUUCGAGCACCCACA-----AAAUAUGC ....((.((((...((((((.(((...((((-(....----.)))))))).)))))).(((((...(((......)))..))))).-----.)))).)) ( -24.10) >consensus GCCAGUUUAUUGUCUUGUUUUGCUU_UGUCU_GUGCGACAGGCAGGCAGCAAAACAAAGUGGAAAACUCUUUUUCCAGCACCCAAAA____AAAUAUUC ...(((((.(..(.((((((((((..(((((........)))))...)))))))))).)..).)))))............................... (-18.11 = -18.68 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:54 2006