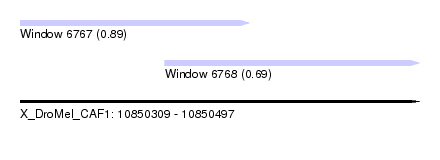
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,850,309 – 10,850,497 |
| Length | 188 |
| Max. P | 0.887986 |

| Location | 10,850,309 – 10,850,417 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -39.69 |
| Consensus MFE | -32.12 |
| Energy contribution | -33.38 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
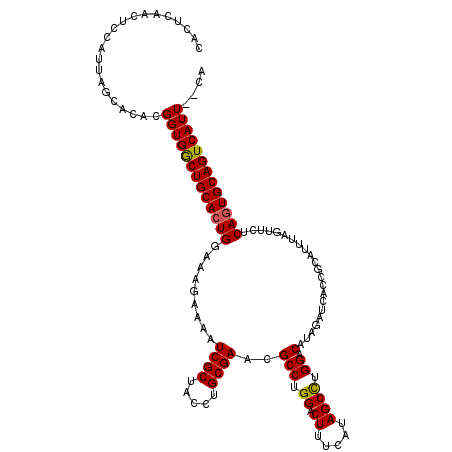
>X_DroMel_CAF1 10850309 108 + 22224390 UUGCUGGCAGACAGGUGGCCACAAGCUUGGCUCUAAAUGCAGAGCACUUGGGGCGCG---CUUGUUAAGGCCACUCAACUCCAUUAGCACACGGUGGCUGCACUGGAAAGA .((((((..((..(((((((((((((...(((((((.(((...))).)))))))..)---)))))...)))))))....))..))))))..(((((....)))))...... ( -41.20) >DroSec_CAF1 4298 110 + 1 -CGUUGGCAGACAGGUGGCCACAAGCUUGGCUCUAAAUGCAGAGCACUUGGGGCACGCUGCUUGUUAAGGCCACUCAACUCCAUUAGCACACGGUGACUGCACUGCAAAGA -.....((((...(((((((((((((.(((((((((.(((...))).))))))).))..))))))...)))))))...........(((...(....)))).))))..... ( -41.00) >DroSim_CAF1 4395 110 + 1 -CGUUGGCAGACAGGUGGCCACAAGCUUGGCUCUAAAUGCAGAGCACUUGGGGCACGCUGCUUGUUAAGGCCACUCAACUCCAUUAGCACACGGUGGCUGCACUGGAAAGA -...(((.((...(((((((((((((.(((((((((.(((...))).))))))).))..))))))...)))))))...)))))........(((((....)))))...... ( -40.40) >DroYak_CAF1 4378 107 + 1 UUGCUGGCAAAUAGGUGGCCACAAGCCUGGCUCUAAAUGCAGAACACUUGGGGCAUG---CUUGUUAAGGCCACUCAACUCCAUUAGCAGAACG-GGCUACACUGGAAAGA (((((((......(((((((((((((...(((((((.((.....)).)))))))..)---)))))...)))))))........)))))))..((-(......)))...... ( -36.14) >consensus _CGCUGGCAGACAGGUGGCCACAAGCUUGGCUCUAAAUGCAGAGCACUUGGGGCACG___CUUGUUAAGGCCACUCAACUCCAUUAGCACACGGUGGCUGCACUGGAAAGA ..(((((.((...(((((((((((((...(((((((.((.....)).))))))).....))))))...)))))))...)))))...))...(((((....)))))...... (-32.12 = -33.38 + 1.25)



| Location | 10,850,377 – 10,850,497 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -28.31 |
| Energy contribution | -28.20 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10850377 120 + 22224390 CACUCAACUCCAUUAGCACACGGUGGCUGCACUGGAAAGAAAUUCGCUACUUGCGAACGCCUGGACUUUCACAGCUUGGGCAUAGAUCACCGCACUUAGCUUUCAGUGCAGUCAUUCUCA .....................((((((((((((((((.((..(((((.....))))).(((..(.((.....)).)..))).....))...((.....)))))))))))))))))).... ( -38.10) >DroSec_CAF1 4368 118 + 1 CACUCAACUCCAUUAGCACACGGUGACUGCACUGCAAAGAAAAUCGCUACCUGCGAACGCCUGGACUUUCAUAGCCUGGACAUAGAUCACCGUAUUUAGUUCUCAGUGCAGUCAUU--CA .....................(((((((((((((...((((..((((.....))))..(((.((.((.....)))).)).)..................)))))))))))))))))--.. ( -32.10) >DroSim_CAF1 4465 118 + 1 CACUCAACUCCAUUAGCACACGGUGGCUGCACUGGAAAGAAAAUCGCUACCUGCGAACGCCUGGACUUUCAUAGCCUGGACAUAGAUCACCGCAUUUAGUUCUCAAUGCAGUCAUU--CA .....................((((((((((.(((((..(((.((((.....))))..(((.((.((.....)))).)).).............)))..))).)).))))))))))--.. ( -28.00) >consensus CACUCAACUCCAUUAGCACACGGUGGCUGCACUGGAAAGAAAAUCGCUACCUGCGAACGCCUGGACUUUCAUAGCCUGGACAUAGAUCACCGCAUUUAGUUCUCAGUGCAGUCAUU__CA .....................(((((((((((((.........((((.....))))..(((.((.((.....)))).)).)......................))))))))))))).... (-28.31 = -28.20 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:48 2006