| Sequence ID | X_DroMel_CAF1 |
|---|---|
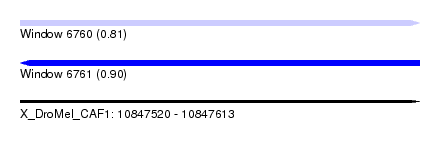
| Location | 10,847,520 – 10,847,613 |
| Length | 93 |
| Max. P | 0.900806 |

| Location | 10,847,520 – 10,847,613 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 94.62 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.66 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10847520 93 + 22224390 GCACUGAGUGUAGUCCCAGGUGAGUCAUGUUGCUAAUUAGUUACGGAUAUGUGCGACAUGCUCACACGAAUACGAAUACACUGAAGCAAGGAU ((..(.((((((.((...((((((.((((((((...................))))))))))))).)......)).)))))).).))...... ( -26.61) >DroSec_CAF1 1556 93 + 1 GUAGUGAGUGUAGUCCCAGGUGAGUCAUGUUGCUAAUUAGUUACGGAUAUGUGCGACAUGCUCACAUGGAUACGAAUACACUGGAGCAAGGAU ((..(.((((((.(((((.(((((.((((((((...................))))))))))))).)))....)).)))))).).))...... ( -29.61) >DroSim_CAF1 1617 93 + 1 GUAGUGAGUGUAGUCCCAGGUGAGUCAUGUUGCUAAUUAGUUACGGAUAUGUGCGACAUGCUCACACGGAUACGAAUACACUGGAGCAAGGAU ((..(.((((((((((...(((((.((((((((...................)))))))))))))..)))).....)))))).).))...... ( -29.11) >DroYak_CAF1 1644 92 + 1 -UAGUGACUGUAGUCCCAGGUGAGUCAUGUUGCUAAUUAGUUACGGAUAUGUGCGACAAGCUCACACGGAUGCGAACACACUGGAGCAAGGAU -(((((..(((.((((...((((((..((((((...................)))))).))))))..))))....)))))))).......... ( -23.91) >consensus GUAGUGAGUGUAGUCCCAGGUGAGUCAUGUUGCUAAUUAGUUACGGAUAUGUGCGACAUGCUCACACGGAUACGAAUACACUGGAGCAAGGAU .(((((..((((.(((...(((((.((((((((...................)))))))))))))..)))))))....))))).......... (-25.28 = -25.66 + 0.38)



| Location | 10,847,520 – 10,847,613 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 94.62 |
| Mean single sequence MFE | -22.31 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.66 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10847520 93 - 22224390 AUCCUUGCUUCAGUGUAUUCGUAUUCGUGUGAGCAUGUCGCACAUAUCCGUAACUAAUUAGCAACAUGACUCACCUGGGACUACACUCAGUGC ......(((..((((((.((.((...(.((((((((((.((...................)).))))).)))))))).)).)))))).))).. ( -22.51) >DroSec_CAF1 1556 93 - 1 AUCCUUGCUCCAGUGUAUUCGUAUCCAUGUGAGCAUGUCGCACAUAUCCGUAACUAAUUAGCAACAUGACUCACCUGGGACUACACUCACUAC ...........((((((...((.((((.((((((((((.((...................)).))))).))))).))))))))))))...... ( -24.11) >DroSim_CAF1 1617 93 - 1 AUCCUUGCUCCAGUGUAUUCGUAUCCGUGUGAGCAUGUCGCACAUAUCCGUAACUAAUUAGCAACAUGACUCACCUGGGACUACACUCACUAC ...........((((((...((.((((.((((((((((.((...................)).))))).))))).))))))))))))...... ( -23.41) >DroYak_CAF1 1644 92 - 1 AUCCUUGCUCCAGUGUGUUCGCAUCCGUGUGAGCUUGUCGCACAUAUCCGUAACUAAUUAGCAACAUGACUCACCUGGGACUACAGUCACUA- .........((((.(((....(((..((((((.....))))))......((..((....))..)))))...)))))))(((....)))....- ( -19.20) >consensus AUCCUUGCUCCAGUGUAUUCGUAUCCGUGUGAGCAUGUCGCACAUAUCCGUAACUAAUUAGCAACAUGACUCACCUGGGACUACACUCACUAC ...........((((((.((....(((.((((((((((.((...................)).))))).))))).))))).))))))...... (-20.72 = -20.66 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:41 2006