
| Sequence ID | X_DroMel_CAF1 |
|---|---|
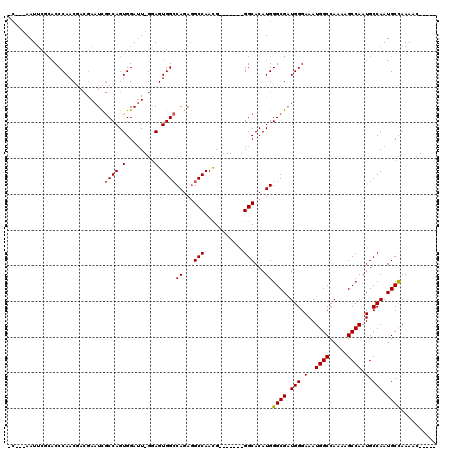
| Location | 10,842,306 – 10,842,492 |
| Length | 186 |
| Max. P | 0.987732 |

| Location | 10,842,306 – 10,842,413 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10842306 107 + 22224390 CCAACAAUUCACACCCAACGACGAAUUGCCAGUGGAUU-GAAGUGGCCAGAAGCCAACG-------GGCACAUGGGCGAUGGGAAAUGGCCAAAAGCCAAUGCCAAUGCCAAAAC----- ((..((((((((...(((.......)))...)))))))-)...((((.....))))..)-------).......((((.(((.(..((((.....)))).).))).)))).....----- ( -29.50) >DroSec_CAF1 21078 97 + 1 -----AAU-----CCCAACGACGAAUCGCCAGUGGAUU-GGAGUGGCCAGAGGCCAACG-------GGCACAUGGGCGAUGGGAAAUGGCCAAAAGCCAAUGCCAAUGCCAAACC----- -----..(-----((((........(((((.(((..((-(...(((((...))))).))-------)...))).))))))))))..((((.....((....))....))))....----- ( -31.30) >DroSim_CAF1 1969 104 + 1 -CA--AAUUCGCACCCAACGACAAAUCGCCAGUGGAUU-GGAGUGGCCAGAGGCCAACG-------GGCACAUGGGCGAUGGGAAAUGGCCAAAAGCCAAUGCCAAUGCCAAAAC----- -..--.(((.(((((((........(((((.(((..((-(...(((((...))))).))-------)...))).)))))))))...((((.....)))).))).)))........----- ( -32.20) >DroYak_CAF1 10076 114 + 1 -C--CAAUUCGCACCCAAC---GAAUCACCAGUGGAAUGGGAGUGGACAGAGGCCAAUGGCCCAGAGGCACAUGGGCGCUGGGAAAUGGCCAAAAGCCAAUGCCAAUGCUAAAGCCAAAG -.--...(((((.((((..---...(((....)))..)))).)))))....(((.....(((((........)))))((..((...((((.....))))...))...))....))).... ( -34.30) >consensus _C___AAUUCGCACCCAACGACGAAUCGCCAGUGGAUU_GGAGUGGCCAGAGGCCAACG_______GGCACAUGGGCGAUGGGAAAUGGCCAAAAGCCAAUGCCAAUGCCAAAAC_____ ...........................((((.(........).))))((...(((...........)))...))((((.(((.(..((((.....)))).).))).)))).......... (-22.54 = -22.60 + 0.06)


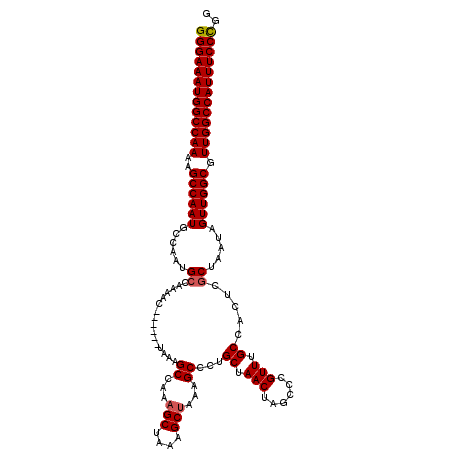
| Location | 10,842,378 – 10,842,492 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -37.81 |
| Energy contribution | -38.12 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10842378 114 + 22224390 GGGAAAUGGCCAAAAGCCAAUGCCAAUGCCAAAAC------UAAAGCCAAAGCUAGAGCUAAAGCCCAGCUAACUAGCCCGUUUGCCACUCGCUAAUAGUUGGCGUUGGCCAUUUCCUGG (((((((((((((..((((((......((......------..((((....(((((((((.......))))..)))))..)))).......)).....)))))).))))))))))))).. ( -41.56) >DroSec_CAF1 21140 114 + 1 GGGAAAUGGCCAAAAGCCAAUGCCAAUGCCAAACC------AAAGGCCAAAGCUAAAGCUAAAGCCCUGCUAACUAGCCCGUUUGCCACUCACUAAUAGUUGGCGUUGGCCAUUUCCCGG (((((((((((((..((((((((....))(((((.------...(((...(((....)))...)))..(((....)))..))))).............)))))).))))))))))))).. ( -43.70) >DroSim_CAF1 2038 114 + 1 GGGAAAUGGCCAAAAGCCAAUGCCAAUGCCAAAAC------UAAAGCCAAAGCUAGAGCUAAAGCCCUGCUAACUAGCCCGUUUGCCACUCGCUAAUAGUUGGCGUUGGCCAUUUCCCGG (((((((((((((..((((((......((......------..((((....((((((((.........)))..)))))..)))).......)).....)))))).))))))))))))).. ( -43.36) >DroYak_CAF1 10150 120 + 1 GGGAAAUGGCCAAAAGCCAAUGCCAAUGCUAAAGCCAAAGCCAAAGCCAAAGCCAAAGCCAAAGCCCUGCUAACUAGCCCGUUUGCCACUCGCUAAUAGUUGGCGUUGGCCAUUUCCCGG (((((((((((((..((((((......(((...((....))...)))...(((....((..((((...(((....)))..)))))).....)))....)))))).))))))))))))).. ( -40.90) >consensus GGGAAAUGGCCAAAAGCCAAUGCCAAUGCCAAAAC______UAAAGCCAAAGCUAAAGCUAAAGCCCUGCUAACUAGCCCGUUUGCCACUCGCUAAUAGUUGGCGUUGGCCAUUUCCCGG (((((((((((((..((((((......((................((...(((....)))...))...((.(((......))).)).....)).....)))))).))))))))))))).. (-37.81 = -38.12 + 0.31)

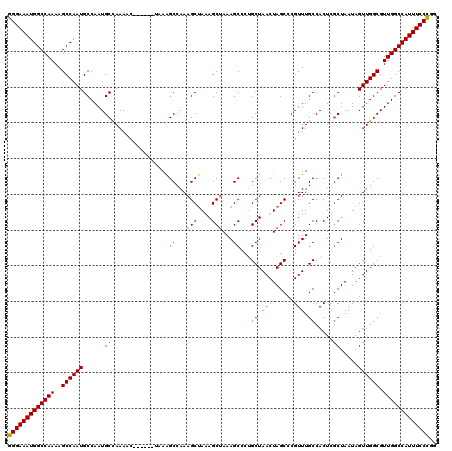
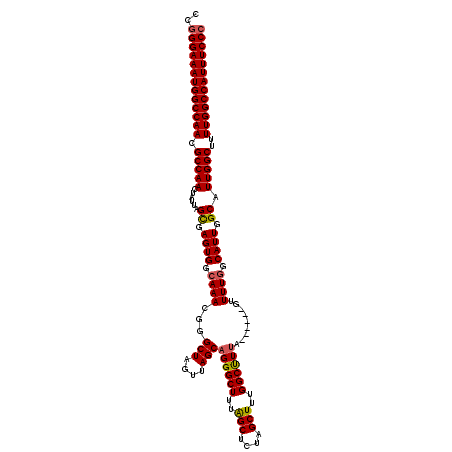
| Location | 10,842,378 – 10,842,492 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -54.15 |
| Consensus MFE | -47.61 |
| Energy contribution | -47.80 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -5.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10842378 114 - 22224390 CCAGGAAAUGGCCAACGCCAACUAUUAGCGAGUGGCAAACGGGCUAGUUAGCUGGGCUUUAGCUCUAGCUUUGGCUUUA------GUUUUGGCAUUGGCAUUGGCUUUUGGCCAUUUCCC ...((((((((((((.(((((......((.((((.(((((.((((((..((((((((....)).))))))))))))...------).)))).)))).)).)))))..)))))))))))). ( -51.70) >DroSec_CAF1 21140 114 - 1 CCGGGAAAUGGCCAACGCCAACUAUUAGUGAGUGGCAAACGGGCUAGUUAGCAGGGCUUUAGCUUUAGCUUUGGCCUUU------GGUUUGGCAUUGGCAUUGGCUUUUGGCCAUUUCCC ..(((((((((((((.(((((......((.((((.((((((((((((..((((((((....))))).))))))))))..------.))))).)))).)).)))))..))))))))))))) ( -55.30) >DroSim_CAF1 2038 114 - 1 CCGGGAAAUGGCCAACGCCAACUAUUAGCGAGUGGCAAACGGGCUAGUUAGCAGGGCUUUAGCUCUAGCUUUGGCUUUA------GUUUUGGCAUUGGCAUUGGCUUUUGGCCAUUUCCC ..(((((((((((((.(((((......((.((((.(((((.((((((..((((((((....))))).)))))))))...------).)))).)))).)).)))))..))))))))))))) ( -53.60) >DroYak_CAF1 10150 120 - 1 CCGGGAAAUGGCCAACGCCAACUAUUAGCGAGUGGCAAACGGGCUAGUUAGCAGGGCUUUGGCUUUGGCUUUGGCUUUGGCUUUGGCUUUAGCAUUGGCAUUGGCUUUUGGCCAUUUCCC ..(((((((((((((.((((((..((.((.....)).))..)((((((..((((((((..((((..(((....)))..))))..)))))).)))))))).)))))..))))))))))))) ( -56.00) >consensus CCGGGAAAUGGCCAACGCCAACUAUUAGCGAGUGGCAAACGGGCUAGUUAGCAGGGCUUUAGCUCUAGCUUUGGCUUUA______GUUUUGGCAUUGGCAUUGGCUUUUGGCCAUUUCCC ..(((((((((((((.(((((......((.((((.((((...(((....)))((((((..(((....)))..)))))).........)))).)))).)).)))))..))))))))))))) (-47.61 = -47.80 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:34 2006