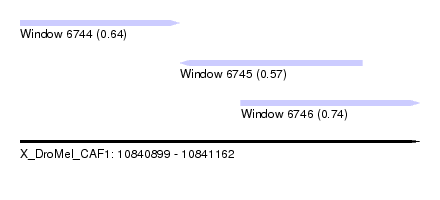
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,840,899 – 10,841,162 |
| Length | 263 |
| Max. P | 0.736525 |

| Location | 10,840,899 – 10,841,004 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -20.59 |
| Energy contribution | -21.37 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10840899 105 + 22224390 CAAACCGAACACC------AGCCCACCGC-------CCACCACCCGCCAACCUCUACUCUAAUGUCCUACCCCCGGGGAGACAUUCUCCCGUGGGUCUUGUGGAGUCAACCCUUUGUU .....((((....------.((((((.((-------((((...............((......))..........(((((.....)))))))))))...)))).))......)))).. ( -24.00) >DroSec_CAF1 19697 118 + 1 CAAACCGAACACCACACACAGCCCACCACCCACUUCCCACCACCCUCCAACCUCUACUCUAAUGUCUCACCCCCGGGGAGACAUUUUCCCGUGGGUCUUGUGGAGUCAACCCUUUGUU ...............(((.((......((((((..(((((....................((((((((.((....)))))))))).....)))))....)))).)).....)).))). ( -26.21) >DroSim_CAF1 569 118 + 1 CAAACCGAACACCACACACAGCCCACCACCCACUUCCCACCACCCGCCAACCUCUACUCUAAUGUCCCACCCCCGGGGAGACAUUUUCCCGUGGGUCUUGUGGAGUCAACCCUUUGUU ......((...(((((....((((((.............................((......)).........((((((....))))))))))))..)))))..))........... ( -23.00) >consensus CAAACCGAACACCACACACAGCCCACCACCCACUUCCCACCACCCGCCAACCUCUACUCUAAUGUCCCACCCCCGGGGAGACAUUUUCCCGUGGGUCUUGUGGAGUCAACCCUUUGUU ......((...(((((....((((((.............................((......))..........((((((...))))))))))))..)))))..))........... (-20.59 = -21.37 + 0.78)



| Location | 10,841,004 – 10,841,124 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -31.49 |
| Energy contribution | -31.30 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10841004 120 - 22224390 AUGCAUAAAUAAAAAUGUGGCAUGUUGGGUCAGAAAUAGCAUGACAUUUUGCCAGCCGGCCGACACAAAAUGGUGGCAGAUACACAGCAAUGGCCAGCGGUAGCAUAAAAUGAAAUGUUC ((((.......(((((((..(((((((.........))))))))))))))(((.((.(((((.(((......)))((.........))..))))).))))).)))).............. ( -30.80) >DroSec_CAF1 19815 120 - 1 AUGCAUAAAUAAAAAUGUGGCAUGUUGGGUCAGAAAUAGCAUGACAUUUUGCCAGCCGGCCGACACAAAAUGGUGGCAGAUACACAGCAGUGGCCAGCGGUAGCAUAAAAUGAAAUGUUC ((((.......(((((((..(((((((.........))))))))))))))(((.((.(((((.(((......)))((.........))..))))).))))).)))).............. ( -30.70) >DroSim_CAF1 687 120 - 1 AUGCAUAAAUAAAAAUGUGGCAUGUUGGGUCAGAAAUAGCAUGACAUUUUGCCAGCCGGCCGACACAAAAUGGUGGCAGAUACACAGCAAUGGCCAGCGGUAGCAUAAAAUGAAAUGUUC ((((.......(((((((..(((((((.........))))))))))))))(((.((.(((((.(((......)))((.........))..))))).))))).)))).............. ( -30.80) >DroYak_CAF1 8798 120 - 1 AUGCAUAAAUAAAAAUGUGGCAUGCUGGGUCAGAAAUAGCAUGACAUUUUGCCAGCCGGCCGACACAAAAUGGUGGCAGACACACAGCAUUGGCCAGCGGUGGCAUAAAAUGAAAUGUUC ((((.......(((((((..(((((((.........))))))))))))))(((.((.(((((((((......)))((.........)).)))))).))))).)))).............. ( -34.00) >consensus AUGCAUAAAUAAAAAUGUGGCAUGUUGGGUCAGAAAUAGCAUGACAUUUUGCCAGCCGGCCGACACAAAAUGGUGGCAGAUACACAGCAAUGGCCAGCGGUAGCAUAAAAUGAAAUGUUC ((((.......(((((((..(((((((.........))))))))))))))(((.((.(((((.(((......)))((.........))..))))).))))).)))).............. (-31.49 = -31.30 + -0.19)



| Location | 10,841,044 – 10,841,162 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -25.09 |
| Consensus MFE | -23.00 |
| Energy contribution | -22.81 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10841044 118 + 22224390 UCUGCCACCAUUUUGUGUCGGCCGGCUGGCAAAAUGUCAUGCUAUUUCUGACCCAACAUGCCACAUUUUUAUUUAUGCAUGCUUCGGCACCCCGGAACCCCUAAACA--UAAAAAUCCCC ........(((.(((.(((((..(((((((.....)))).)))....))))).))).)))...........((((((..(((....)))....((....))....))--))))....... ( -24.10) >DroSec_CAF1 19855 118 + 1 UCUGCCACCAUUUUGUGUCGGCCGGCUGGCAAAAUGUCAUGCUAUUUCUGACCCAACAUGCCACAUUUUUAUUUAUGCAUGCUUCGGCACCCAGGAACCCCUAAACA--UAAAAAUCCCC ........(((.(((.(((((..(((((((.....)))).)))....))))).))).)))...........((((((..(((....)))...(((....)))...))--))))....... ( -25.20) >DroSim_CAF1 727 118 + 1 UCUGCCACCAUUUUGUGUCGGCCGGCUGGCAAAAUGUCAUGCUAUUUCUGACCCAACAUGCCACAUUUUUAUUUAUGCAUGCUUCGGCACCCAGGAACCCCUAAACA--UAAAAAUCUCC ........(((.(((.(((((..(((((((.....)))).)))....))))).))).)))...........((((((..(((....)))...(((....)))...))--))))....... ( -25.20) >DroYak_CAF1 8838 120 + 1 UCUGCCACCAUUUUGUGUCGGCCGGCUGGCAAAAUGUCAUGCUAUUUCUGACCCAGCAUGCCACAUUUUUAUUUAUGCAUGCCUCGGCACCCAGAAACCCCUAAACCCCCAAAAACCCCU ..((((.......((.(((((..(((((((.....)))).)))....))))).))((((((...............))))))...))))............................... ( -25.86) >consensus UCUGCCACCAUUUUGUGUCGGCCGGCUGGCAAAAUGUCAUGCUAUUUCUGACCCAACAUGCCACAUUUUUAUUUAUGCAUGCUUCGGCACCCAGGAACCCCUAAACA__UAAAAAUCCCC ..((((......(((.(((((..(((((((.....)))).)))....))))).)))(((((...............)))))....))))............................... (-23.00 = -22.81 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:27 2006