
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,830,881 – 10,831,019 |
| Length | 138 |
| Max. P | 0.962244 |

| Location | 10,830,881 – 10,830,998 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -30.88 |
| Energy contribution | -31.63 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10830881 117 + 22224390 UGCGGACUGUUUUCCGGUCCCGCCAAAUAGCAUUUCCUCGAUGCCAGUGCAGCACUGAUUCAACGGCACUGCGCAGCUUGUUGUGGCAAUAAAUGGCAUUAUGCUGCUUAGCAAUCA .(.((((((.....)))))))((.((.((((((......(((((((((((((..(((......)))..)))))).(((......)))......))))))))))))).)).))..... ( -39.50) >DroSim_CAF1 36989 117 + 1 UGCGGACUGUUUUCCGGUCCCGCCAAAGAGCAUUUGCUCGAUGCCAGUGCAGCACUGAUUCAACGGCACUGUGCAGCUUGUUGUGGCAAUAAAUGGCAUUAUGCUGCUUAGCAAUCA .((((((((.....)))).))))....((((....))))((((((((..(((..(((......)))..)))..).(((......)))......))))))).((((....)))).... ( -40.20) >DroEre_CAF1 54050 100 + 1 UGCGGACUGCUUUCCGGUGCCGCCAAG-----------------CAGUGCAGCACUGAUUCCACGGCACUGUGCAGCUUGUUGUGGCAAUAAAUGGCAUUAUGCUGCUUAGCAAUCA ....((.((((...((((((((.....-----------------(((((...)))))......)))))))).(((((.(((..(........)..)))....)))))..)))).)). ( -36.30) >consensus UGCGGACUGUUUUCCGGUCCCGCCAAA_AGCAUUU_CUCGAUGCCAGUGCAGCACUGAUUCAACGGCACUGUGCAGCUUGUUGUGGCAAUAAAUGGCAUUAUGCUGCUUAGCAAUCA (((((((((.....)))))).(((.....(((((.....)))))(((((...))))).......))).....(((((.(((..(........)..)))....)))))...))).... (-30.88 = -31.63 + 0.75)


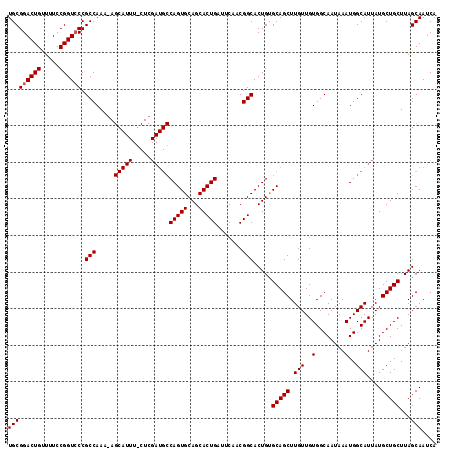
| Location | 10,830,921 – 10,831,019 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -28.49 |
| Energy contribution | -28.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10830921 98 + 22224390 AUGCCAGUGCAGCACUGAUUCAACGGCACUGCGCAGCUUGUUGUGGCAAUAAAUGGCAUUAUGCUGCUUAGCAAUCAUGCCACUUGAUUGUUGUGGUG ((((((((((((..(((......)))..)))))).(((......)))......))))))...((..(..((((((((.......)))))))))..)). ( -32.90) >DroSim_CAF1 37029 98 + 1 AUGCCAGUGCAGCACUGAUUCAACGGCACUGUGCAGCUUGUUGUGGCAAUAAAUGGCAUUAUGCUGCUUAGCAAUCAUGCCACUUGAUUGUUGUGGUG (((((((..(((..(((......)))..)))..).(((......)))......))))))...((..(..((((((((.......)))))))))..)). ( -30.50) >DroEre_CAF1 54077 94 + 1 ----CAGUGCAGCACUGAUUCCACGGCACUGUGCAGCUUGUUGUGGCAAUAAAUGGCAUUAUGCUGCUUAGCAAUCAUGCCACUUGAUUGUUGUGGUG ----(((((...)))))...(((((((.(((.(((((.(((..(........)..)))....))))).))).(((((.......)))))))))))).. ( -30.10) >consensus AUGCCAGUGCAGCACUGAUUCAACGGCACUGUGCAGCUUGUUGUGGCAAUAAAUGGCAUUAUGCUGCUUAGCAAUCAUGCCACUUGAUUGUUGUGGUG ....(((((...))))).........(((..((((((.(((..(........)..)))....)))))..((((((((.......)))))))))..))) (-28.49 = -28.27 + -0.22)



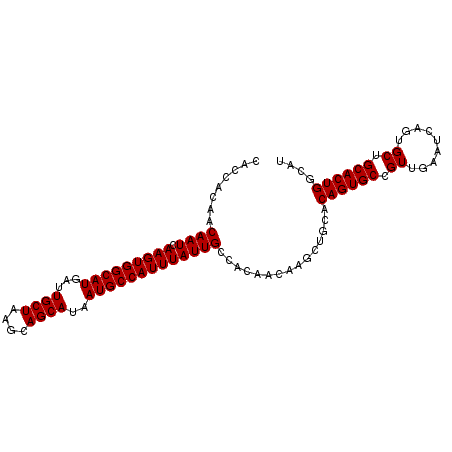
| Location | 10,830,921 – 10,831,019 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -27.43 |
| Energy contribution | -27.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10830921 98 - 22224390 CACCACAACAAUCAAGUGGCAUGAUUGCUAAGCAGCAUAAUGCCAUUUAUUGCCACAACAAGCUGCGCAGUGCCGUUGAAUCAGUGCUGCACUGGCAU ........((((.(((((((((...((((....))))..))))))))))))).........((((.((((..(.(......).)..)))).).))).. ( -31.40) >DroSim_CAF1 37029 98 - 1 CACCACAACAAUCAAGUGGCAUGAUUGCUAAGCAGCAUAAUGCCAUUUAUUGCCACAACAAGCUGCACAGUGCCGUUGAAUCAGUGCUGCACUGGCAU ........((((.(((((((((...((((....))))..)))))))))))))...........(((.((((((.((.........)).))))))))). ( -31.40) >DroEre_CAF1 54077 94 - 1 CACCACAACAAUCAAGUGGCAUGAUUGCUAAGCAGCAUAAUGCCAUUUAUUGCCACAACAAGCUGCACAGUGCCGUGGAAUCAGUGCUGCACUG---- ..((((..((((.(((((((((...((((....))))..))))))))))))).........((........)).))))...(((((...)))))---- ( -28.10) >consensus CACCACAACAAUCAAGUGGCAUGAUUGCUAAGCAGCAUAAUGCCAUUUAUUGCCACAACAAGCUGCACAGUGCCGUUGAAUCAGUGCUGCACUGGCAU ........((((.(((((((((...((((....))))..)))))))))))))...............((((((.((.........)).)))))).... (-27.43 = -27.43 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:22 2006