| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,811,478 – 10,811,575 |
| Length | 97 |
| Max. P | 0.500000 |

| Location | 10,811,478 – 10,811,575 |
|---|---|
| Length | 97 |
| Sequences | 6 |
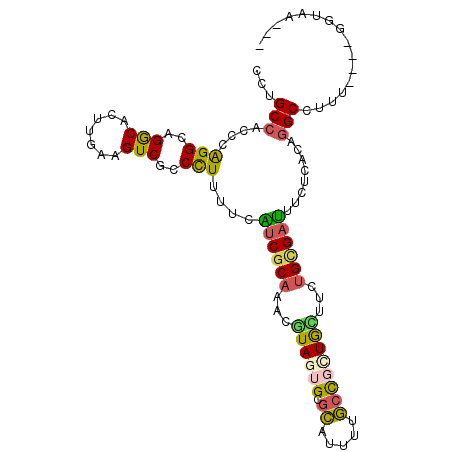
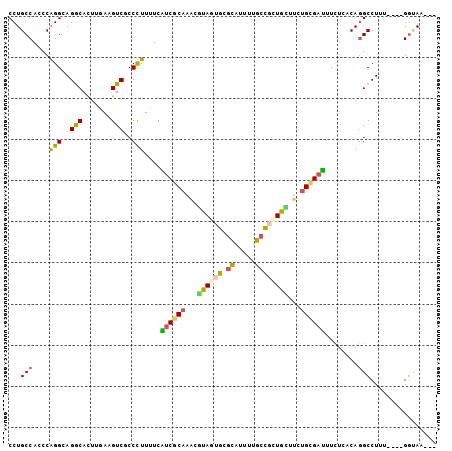
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -17.32 |
| Energy contribution | -16.63 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10811478 97 + 22224390 CCUGCCACCCAGGAAGGCACUUGAAGUCACCUUUUUCAUCGCAAACAUAGUGUGCAUUUUGCCGCUGUUUCUGUGAUUUCUCACAGGCCUUA----GGUAA--- ((((.....))))(((((...(((((.......)))))........((((((.((.....))))))))..((((((....))))))))))).----.....--- ( -26.60) >DroVir_CAF1 24827 97 + 1 CCUGCCAGCCAGGCAGGCAUUUCACGUCGCCCUUUUCGUCGCAGGUGUAGUGCGAGUUCAUCUUGUACUUUUGCGUCUUUUCGCAGGCCUGU----GGCAA--- ((((((.....))))))....................(((((((((..((((((((.....)))))))).(((((......)))))))))))----)))..--- ( -39.80) >DroEre_CAF1 31702 96 + 1 CCUGCCAGCCAGGAAGACACUUAAAGUCGCCCUUUUCAUCGCAAAUGUAGUGUGCGUUUUGCCGCUGCUUCUGCGAUUUCUCACAGGCCUUA----GG-AU--- ((((...((((((..(((.......)))..)))....((((((...((((((.((.....))))))))...))))))........)))..))----))-..--- ( -30.50) >DroYak_CAF1 30533 97 + 1 CCUGCCACCCAGGCAGACACUUGAAGUCGCCCUUUUCAUCGCAAACAUAGUGUGCAUUUUGCCGCUGCUUCUGCGAUUUCUCACACGCCUUG----GGUAA--- ......((((((((.(((.......))).........((((((....(((((.((.....)))))))....)))))).........))).))----)))..--- ( -29.60) >DroMoj_CAF1 28774 100 + 1 CCUGCCAGCCGGGCAGGCACUUCACGUCCCCCUUCUCGUCGCAGCUGUAGUGCGAGUUCAUCUGGUACUUGGGCGACUUCUCGCAGGCCUGU----GCCAAUGG ((((((.....)))))).................((((.(((.......)))))))..(((.((((((..((((((....))))...)).))----))))))). ( -31.60) >DroAna_CAF1 26457 101 + 1 CCUGCCACCCAGGGAGGCACUUGAAGUCGCCCUUCUCGUCGCAGACGUAGUGCACACUUUGCUUUUGUUUUUGGGAUUUCUCACAGGCCUAUGAAUGGUAA--- ..(((((..((...((((.((.((((((((.((...((((...)))).)).))...((..((....))....))))))))....)))))).))..))))).--- ( -25.90) >consensus CCUGCCACCCAGGCAGGCACUUGAAGUCGCCCUUUUCAUCGCAAACGUAGUGCGCAUUUUGCCGCUGCUUCUGCGAUUUCUCACAGGCCUUU____GGUAA___ ...(((....(((..(((.......)))..)))....((((((...((((((.((.....))))))))...))))))........)))................ (-17.32 = -16.63 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:17 2006