
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,805,010 – 10,805,174 |
| Length | 164 |
| Max. P | 0.988236 |

| Location | 10,805,010 – 10,805,115 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.29 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -14.24 |
| Energy contribution | -15.72 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
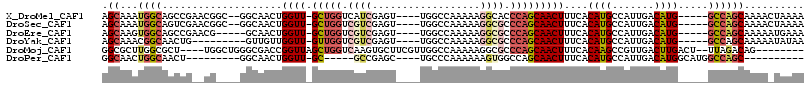
>X_DroMel_CAF1 10805010 105 + 22224390 AGCAAAUGGCAGCCGAACGGC--GGCAACUGGUU-GCUGGUCAUCGAGU----UGGCCAAAAAGGCACCCAGCAACUUUCACAUGCCAUUGACAUG-----GCCAGCAAAACUAAAA .((...((((.(((....)))--((((...((((-(((((.((......----))(((.....)))..)))))))))......)))).........-----)))))).......... ( -36.70) >DroSec_CAF1 20259 105 + 1 AGCAAAUGGCAGUCGAACGGC--GGCAACUGGUU-GCUGGUCGUCGAGU----UGGCCAAAAAGGCGCCCAGCAACUUUCACAUGCCAUUGACAUG-----GCCAGCAAAACUAAAA .((...((((.(((((..(((--(......((((-(((((.((((...(----(....))...)))).)))))))))......)))).)))))...-----)))))).......... ( -39.10) >DroEre_CAF1 25633 102 + 1 AGCAAGUGGCAGCCGAACG-----GCAACUGGUU-GCUGGUCGUCGAGU----UGGCCAAAAAGGCGCCCAGCAACUUUCACAUGCCAUUGACAUG-----GCCAGCAAAAAUGAAA .((..((((..(((....)-----))....((((-(((((.((((...(----(....))...)))).))))))))).))))..(((((....)))-----))..)).......... ( -36.10) >DroYak_CAF1 24278 98 + 1 AGCAAACGGCAACUG---------GUUGUUGGUU-GUUGGUCGUCGAGU----UGGCCAAAAAGGCGCCCAGCAACUUUCACAUGCCAUUGACAUG-----GCCAGCAAAAAUAUAA .((.....))..(((---------((((..((((-(((((.((((...(----(....))...)))).)))))))))..))((((.......))))-----)))))........... ( -29.50) >DroMoj_CAF1 22053 103 + 1 GGCGCUUGGCGCU----UGGCUGGGCGACCGGUUAGCUGGUCAAGUGCUUCGUUGGCCAAAAAGGCGCCCAGCAACUUUCACAAGCCGUUGACUUGACU--UUAGACAG-------- ((((((((.....----..(((((((((((((....)))))).............(((.....))))))))))........)))).)))).........--........-------- ( -38.36) >DroPer_CAF1 22426 88 + 1 GGCAACUGGCAACU---------GGCAACUGGUU-GC-----GCCGAGC----UGCCCAAAAAAGUGGCCAGCAACUUUCACAUGCCAUUGACAUGGCAUGGCCAGC---------- (((.....((((((---------(....).))))-))-----.....((----((.(((......))).))))........((((((((....)))))))))))...---------- ( -35.30) >consensus AGCAAAUGGCAGCCG__CG____GGCAACUGGUU_GCUGGUCGUCGAGU____UGGCCAAAAAGGCGCCCAGCAACUUUCACAUGCCAUUGACAUG_____GCCAGCAAAA_UAAAA .((...((((....................((((.(((((.((((..................)))).)))))))))....((((.......)))).....)))))).......... (-14.24 = -15.72 + 1.48)

| Location | 10,805,080 – 10,805,174 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.52 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.556137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10805080 94 - 22224390 UUGCACCGUGCAACUUGCAACUUGCA--------GCCCGCCAAUCGCAUAUAUAUUGUAUUUUUUUUUUUUAGUUUUGCUGGCCAUGUCAAUGGCAUGUGAA (((((...(((.....)))...))))--------)........((((((.....................(((.....)))(((((....))))))))))). ( -21.20) >DroSec_CAF1 20329 91 - 1 UUGCAGCGUGCAACUUGCAACUUGCA--------GCCCGCCAAUCGCAUAUAUAUU---UUUUUUCUUUUUAGUUUUGCUGGCCAUGUCAAUGGCAUGUGAA ((((((..(((.....)))..)))))--------)........((((((.......---...........(((.....)))(((((....))))))))))). ( -21.90) >DroEre_CAF1 25700 86 - 1 UUGCACCGUGCAACUCGCAACUUGCAACUC-GCUGCCCGCCACGCGUAUAU---------------UUUUCAUUUUUGCUGGCCAUGUCAAUGGCAUGUGAA (((((...(((.....)))...))))).((-((((((.((((.(((.....---------------..........))))))).((....)))))).)))). ( -22.16) >DroYak_CAF1 24341 87 - 1 UUGCAGCGUGCAACUUGCAACUUGCAACGCGGCUGCCCGCCACGCGCAUAU---------------UUUAUAUUUUUGCUGGCCAUGUCAAUGGCAUGUGAA .(((.(((((....((((.....)))).((((....))))))))))))...---------------........((..(..(((((....)))))..)..)) ( -27.50) >consensus UUGCACCGUGCAACUUGCAACUUGCA________GCCCGCCAAGCGCAUAU_______________UUUUUAGUUUUGCUGGCCAUGUCAAUGGCAUGUGAA .((((...(((.....)))...))))............((((...(((............................)))))))(((((.....))))).... (-16.70 = -16.52 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:14 2006