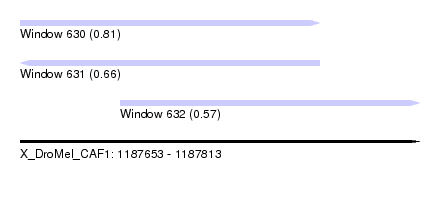
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,187,653 – 1,187,813 |
| Length | 160 |
| Max. P | 0.810062 |

| Location | 1,187,653 – 1,187,773 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -51.30 |
| Consensus MFE | -42.79 |
| Energy contribution | -43.10 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1187653 120 + 22224390 UGCAGCUGGCUUUCGAGCUGUUGCGACAGCUCGUGGGGCAGAGCGACGCCUAGCUUCUGCAACUGUUGGGUAUUGUAGAGCAGUUUCUGCAGCUCCUCCAGAUCCCCCUUUCGGGGCCCA ((((((((.((..((((((((....))))))))..)).)))(((........))).))))).(((..(((..((((((((....))))))))..))).)))...((((....)))).... ( -48.80) >DroSec_CAF1 23530 120 + 1 UGCAGCUGGCUUUCGAGCUGCUGCGACAGCUCGUGGGGCAGAGCGACGCCUAACUUCUGCAGCUGUUGGGUGUUGUAGAGCAGUUUCUGCAGCUCCUCCAGAUCCGCCUUUCGGGGUCCC .(((((.((((....)))))))))(((..((((..((((.((((((((((((((..(....)..)))))))))))).((((.((....)).)))).......)).))))..))))))).. ( -51.00) >DroEre_CAF1 22880 120 + 1 UGCAGCUGGCUUUCGAGCUGCUGCGACAACUCGUGGGGCAGAGCAACGCCUAGCUUCUGCAGCUGUUGGGUGUUGUAGAGCAGUUUCUGCAGCUCCUCCAGAUCCGCCUUUCUGGGCCCA .(((((.((((....)))))))))...........((((.((((((((((((((..(....)..))))))))))((((((....)))))).))))..(((((........))))))))). ( -52.30) >DroAna_CAF1 8135 120 + 1 UGCAGCUGGCUCUCCAAUUGCUGGGAGAGCUCGUGGGGCAGGGCCACGCCCAGCUUCUGCAGCUGCUGGGUGUUGUAGAGCAGCUUCUGAAGCUCCUCCAGAUCCGCCUUCCGGGGACCC .((((((((((((((........)))))))....(((((.((((...)))).)))))..))))))).((((......((((..(....)..))))((((.((.......)).)))))))) ( -53.10) >consensus UGCAGCUGGCUUUCGAGCUGCUGCGACAGCUCGUGGGGCAGAGCGACGCCUAGCUUCUGCAGCUGUUGGGUGUUGUAGAGCAGUUUCUGCAGCUCCUCCAGAUCCGCCUUUCGGGGCCCA ((((((.((((....))))))))))....((((..((((.((((((((((((((..........)))))))))))).((((.((....)).)))).......)).))))..))))..... (-42.79 = -43.10 + 0.31)

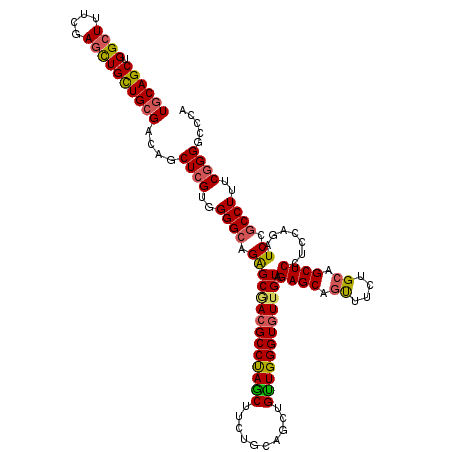
| Location | 1,187,653 – 1,187,773 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -49.10 |
| Consensus MFE | -34.25 |
| Energy contribution | -34.88 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1187653 120 - 22224390 UGGGCCCCGAAAGGGGGAUCUGGAGGAGCUGCAGAAACUGCUCUACAAUACCCAACAGUUGCAGAAGCUAGGCGUCGCUCUGCCCCACGAGCUGUCGCAACAGCUCGAAAGCCAGCUGCA ((((((((....))))....((..(((((..........))))).))...)))).....(((((..(((.((((......))))...((((((((....))))))))..)))...))))) ( -49.00) >DroSec_CAF1 23530 120 - 1 GGGACCCCGAAAGGCGGAUCUGGAGGAGCUGCAGAAACUGCUCUACAACACCCAACAGCUGCAGAAGUUAGGCGUCGCUCUGCCCCACGAGCUGUCGCAGCAGCUCGAAAGCCAGCUGCA (((.((((....)).))...((..(((((..........))))).))...)))..(((((((........((((......))))...((((((((....))))))))...).)))))).. ( -45.60) >DroEre_CAF1 22880 120 - 1 UGGGCCCAGAAAGGCGGAUCUGGAGGAGCUGCAGAAACUGCUCUACAACACCCAACAGCUGCAGAAGCUAGGCGUUGCUCUGCCCCACGAGUUGUCGCAGCAGCUCGAAAGCCAGCUGCA ((((.(((((........))))).(((((..........)))))......)))).((((((((((.((........)))))))....((((((((....))))))))......))))).. ( -43.50) >DroAna_CAF1 8135 120 - 1 GGGUCCCCGGAAGGCGGAUCUGGAGGAGCUUCAGAAGCUGCUCUACAACACCCAGCAGCUGCAGAAGCUGGGCGUGGCCCUGCCCCACGAGCUCUCCCAGCAAUUGGAGAGCCAGCUGCA ((((((((....)).)))..(((((.((((.....)))).))))).....))).(((((((.....((.((((...)))).)).......(((((((.(....).)))))))))))))). ( -58.30) >consensus GGGGCCCCGAAAGGCGGAUCUGGAGGAGCUGCAGAAACUGCUCUACAACACCCAACAGCUGCAGAAGCUAGGCGUCGCUCUGCCCCACGAGCUGUCGCAGCAGCUCGAAAGCCAGCUGCA (((.((((....)).))...((..(((((..........))))).))...)))..((((((.....(((.((((......))))...((((((((....))))))))..))))))))).. (-34.25 = -34.88 + 0.62)

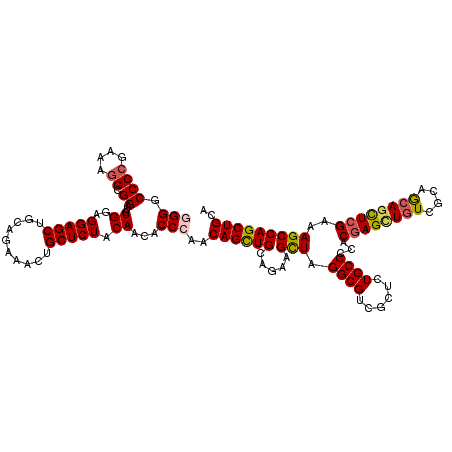
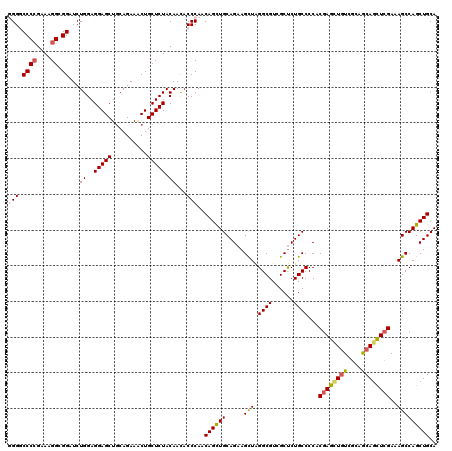
| Location | 1,187,693 – 1,187,813 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -51.78 |
| Consensus MFE | -35.79 |
| Energy contribution | -37.73 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1187693 120 + 22224390 GAGCGACGCCUAGCUUCUGCAACUGUUGGGUAUUGUAGAGCAGUUUCUGCAGCUCCUCCAGAUCCCCCUUUCGGGGCCCAGAGCUGGGCGUAGUGGGAGCCCUCCUUGCGGGCACGAAAC ..((.((((((((((.(((...(((..(((..((((((((....))))))))..))).)))...((((....))))..))))))))))))).))....((((.......))))....... ( -49.80) >DroSec_CAF1 23570 120 + 1 GAGCGACGCCUAACUUCUGCAGCUGUUGGGUGUUGUAGAGCAGUUUCUGCAGCUCCUCCAGAUCCGCCUUUCGGGGUCCCGAGCUGGGCGUGGUGGGAGCCCUCCUUGCGGGCACGAAAC ((((((((((((((..(....)..))))))))))((((((....)))))).)))).(((..(((((((((((((....)))))..))))).))).)))((((.......))))....... ( -51.80) >DroEre_CAF1 22920 120 + 1 GAGCAACGCCUAGCUUCUGCAGCUGUUGGGUGUUGUAGAGCAGUUUCUGCAGCUCCUCCAGAUCCGCCUUUCUGGGCCCAGAGCUGGGCGUAGUGGGUGCCCUCCGCGCGGGCACGAAAC ..((.((((((((((.(((..((....(((.(((((((((....))))))))).)))(((((........))))))).))))))))))))).))..((((((.......))))))..... ( -56.70) >DroAna_CAF1 8175 117 + 1 GGGCCACGCCCAGCUUCUGCAGCUGCUGGGUGUUGUAGAGCAGCUUCUGAAGCUCCUCCAGAUCCGCCUUCCGGGGACCCGAGCUCAGAGGCGU---CACCUUCUUGGAGGGCACAAAGC ..((.(((((((((..(....)..))))))))).))...((.((((((((.((((((((.((.......)).))))....))))))))))))..---..((((....))))))....... ( -48.80) >consensus GAGCGACGCCUAGCUUCUGCAGCUGUUGGGUGUUGUAGAGCAGUUUCUGCAGCUCCUCCAGAUCCGCCUUUCGGGGCCCAGAGCUGGGCGUAGUGGGAGCCCUCCUUGCGGGCACGAAAC ..((.(((((((((((..((..(((..(((.(((((((((....))))))))))))..))).....((.....))))...))))))))))).))....((((.......))))....... (-35.79 = -37.73 + 1.94)

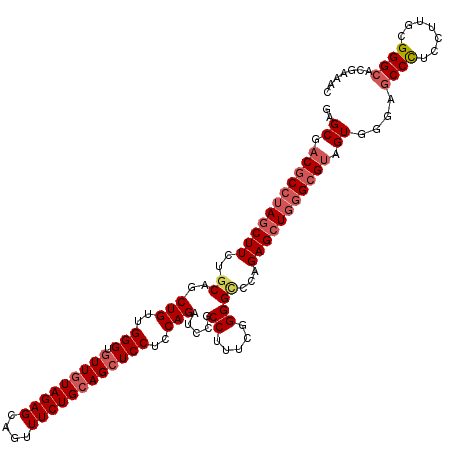
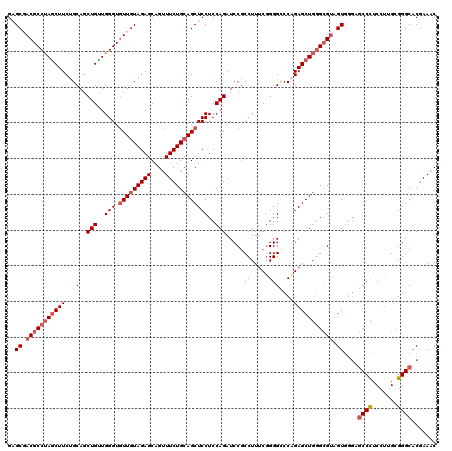
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:57 2006