
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,780,796 – 10,780,980 |
| Length | 184 |
| Max. P | 0.640030 |

| Location | 10,780,796 – 10,780,888 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -19.13 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
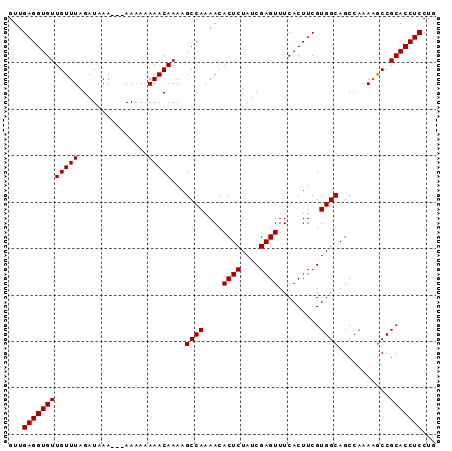
>X_DroMel_CAF1 10780796 92 + 22224390 GUUGAGGUGUUGUUUAGAUUAAAAAAAAAAAAACAAAAGCCAAAACACUCUAUCGAGUUUCACUUCGUGGCAGCCAAAAACCGCACCUCCUG ...((((((((((((...............)))))...((((....((((....)))).........))))...........)))))))... ( -18.48) >DroSec_CAF1 30347 89 + 1 GUUGAGGUGUUGUUUAGAUAAA---AAAAAAAACAAAAGCCAAAACACUCUAUCGAGUUUCACUUCGUGGCAGCCAAACGCCGCACCUCCUG ...((((((((((((.......---.....))))))..........((((....))))..))))))(((((........)))))........ ( -19.40) >DroSim_CAF1 29009 88 + 1 GUUGAGGUGUUGUUUAGAUAAA---AA-AAAAACAAAAGCCAAAACACUCUAUCGAGUUUCACUUCGUGGCAGCCAAAAGCCGCACCUCCUG ...((((((((((((.......---..-..))))))..........((((....))))..))))))(((((........)))))........ ( -19.50) >consensus GUUGAGGUGUUGUUUAGAUAAA___AAAAAAAACAAAAGCCAAAACACUCUAUCGAGUUUCACUUCGUGGCAGCCAAAAGCCGCACCUCCUG ...((((((((((((...............)))))...((((....((((....)))).........))))...........)))))))... (-18.48 = -18.48 + 0.00)


| Location | 10,780,822 – 10,780,928 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 95.60 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -17.89 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
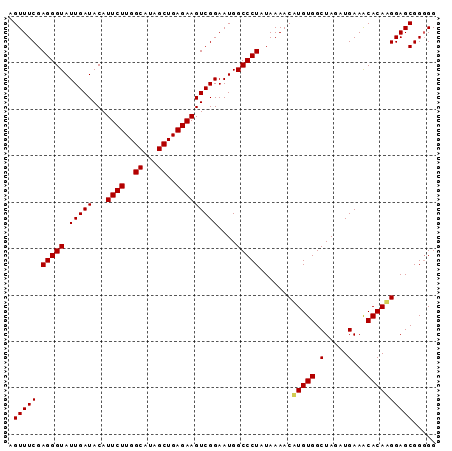
>X_DroMel_CAF1 10780822 106 + 22224390 AAAAAAACAAAAGCCAAAACACUCUAUCGAGUUUCACUUCGUGGCAGCCAAAAACCGCACCUCCUGCCACCGCUCCAUGUGUUUCAUCUAGCCACAUGUUUUAUAG ...........(((......((((....))))........(((((((................))))))).))).((((((((......)).))))))........ ( -19.49) >DroSec_CAF1 30370 106 + 1 AAAAAAACAAAAGCCAAAACACUCUAUCGAGUUUCACUUCGUGGCAGCCAAACGCCGCACCUCCUGCCCCCGCUCCUUGUGUUUCAUCUAGCCACAAGUUUUAUAG ...............(((((((((....))))........(((((.((.....)).((((.....((....)).....))))........)))))..))))).... ( -18.70) >DroSim_CAF1 29032 105 + 1 A-AAAAACAAAAGCCAAAACACUCUAUCGAGUUUCACUUCGUGGCAGCCAAAAGCCGCACCUCCUGCCCCCGCUCCUUGUGCUUCAUCUAGCCACAUGUUUUAUAG .-.((((((...........((((....))))........(((((.((.....)).((((.....((....)).....))))........))))).)))))).... ( -22.10) >consensus AAAAAAACAAAAGCCAAAACACUCUAUCGAGUUUCACUUCGUGGCAGCCAAAAGCCGCACCUCCUGCCCCCGCUCCUUGUGUUUCAUCUAGCCACAUGUUUUAUAG ...............(((((((((....))))........(((((.((.....)).((((.....((....)).....))))........)))))..))))).... (-17.89 = -18.00 + 0.11)



| Location | 10,780,888 – 10,780,980 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 97.10 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -23.95 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
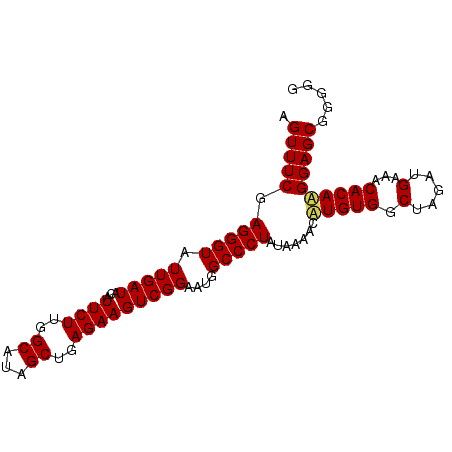
>X_DroMel_CAF1 10780888 92 - 22224390 AGUUUCGAGGGUAUUGAUACAUUCUUGGCAUAGCUGAGAAGUCGGAAUGGCCCUAUAAAACAUGUGGCUAGAUGAAACACAUGGAGCGGUGG .((((..(((((.(((((...((((..((...))..)))))))))....)))))......((((((.(.....)...))))))))))..... ( -25.30) >DroSec_CAF1 30436 92 - 1 AGUUUCGAGGGUAUUGAUACAUUCUUGGCAUAGCUGAGAAGUCGGAAUGGCCCUAUAAAACUUGUGGCUAGAUGAAACACAAGGAGCGGGGG ..(..(((((((.(((((...((((..((...))..)))))))))....)))))......((((((.(.....)...))))))...))..). ( -26.50) >DroSim_CAF1 29097 92 - 1 AGUUUCGAGGGUAUUGAUACAUUCUUGGCAUAGCUGAGAAGUCGGAAUGGCCCUAUAAAACAUGUGGCUAGAUGAAGCACAAGGAGCGGGGG .(((((.(((((.(((((...((((..((...))..)))))))))....)))))........((((.((......)))))).)))))..... ( -25.20) >consensus AGUUUCGAGGGUAUUGAUACAUUCUUGGCAUAGCUGAGAAGUCGGAAUGGCCCUAUAAAACAUGUGGCUAGAUGAAACACAAGGAGCGGGGG .(((((.(((((.(((((...((((..((...))..)))))))))....))))).......(((((.(.....)...))))))))))..... (-23.95 = -24.07 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:58 2006