
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,773,588 – 10,773,710 |
| Length | 122 |
| Max. P | 0.766825 |

| Location | 10,773,588 – 10,773,683 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -24.23 |
| Energy contribution | -24.15 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10773588 95 + 22224390 UCGCCAACGCCU---ACACCGGGAUCUCCUUCGGCUUGGCAAUCACCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCCACUAAGGCUACUUCGGC-- ..(((.......---.(((.(((((...((..(((((((((((....)))))))).)))..)).)))))))).....(((......)))......)))-- ( -30.00) >DroVir_CAF1 27436 97 + 1 ---CCAACGCCAAGCACGCCAACAUCGCCAUCCGCGUGGCAGGCGCCUUUGCCGACGCCUGUGCAUCCAGCUGUGCCGGCUGUUAAGCCAGCUCCGGCAG ---.....(((..((((((((....(((.....)))))))(((((.(......).)))))))))..........((.((((....)))).))...))).. ( -36.70) >DroSec_CAF1 22968 95 + 1 UCGCCAACGCCC---ACACCGGGAUCUCCUUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCAACCAAGGCUACUUCGGC-- ..(((.......---.(((.(((((...((..(((((((((((....)))))))).)))..)).)))))))).....(((......)))......)))-- ( -30.50) >DroSim_CAF1 21518 95 + 1 UCGCCAACGCCC---ACACCGGGAUCUCCUUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCCACGAAGGCUACAUCGGC-- ..(((....(((---.....)))....((((((((((((((((....)))))))).)))...((((...))))..........))))).......)))-- ( -32.10) >DroEre_CAF1 20657 95 + 1 UCGCCAACGCCC---ACACCGGGAUCUCCUUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCCGUGCAUCCUCCCACCAAGGCUACAUCAGC-- ..(((.......---.....(((((...((..(((((((((((....)))))))).)))..)).)))))(((........)))...))).........-- ( -28.20) >DroYak_CAF1 22497 95 + 1 UCGCCUACGCCC---ACGCCGGGCUCUCCAUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCCACCAAGGCUACGUCGGC-- ..(((.(((...---..(((((((........(((((((((((....)))))))).)))...((((...))))....)))).....)))..))).)))-- ( -33.70) >consensus UCGCCAACGCCC___ACACCGGGAUCUCCUUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCCACCAAGGCUACUUCGGC__ ........(((.....(((.(((((((.....(((((((((((....)))))))).)))..)).)))))))).....(((......)))......))).. (-24.23 = -24.15 + -0.08)

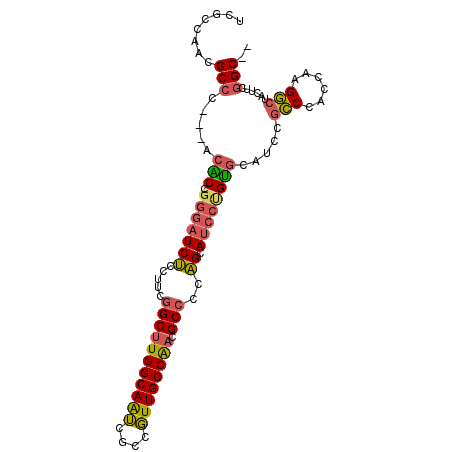
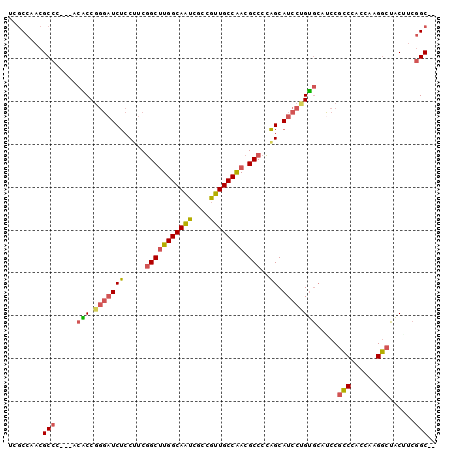
| Location | 10,773,588 – 10,773,683 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -30.52 |
| Energy contribution | -31.38 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10773588 95 - 22224390 --GCCGAAGUAGCCUUAGUGGGCGGAUGCACAGGAUGCUGGGGCGUUGGCAACGGUGAUUGCCAAGCCGAAGGAGAUCCCGGUGU---AGGCGUUGGCGA --(((((.((.((((....))))...(((((.((((.((..(((.(((((((......))))))))))...))..))))..))))---).)).))))).. ( -39.00) >DroVir_CAF1 27436 97 - 1 CUGCCGGAGCUGGCUUAACAGCCGGCACAGCUGGAUGCACAGGCGUCGGCAAAGGCGCCUGCCACGCGGAUGGCGAUGUUGGCGUGCUUGGCGUUGG--- ..(((((.(((((((....)))))))..........(((((((((((......)))))))(((((((.....)))....))))))))))))).....--- ( -45.20) >DroSec_CAF1 22968 95 - 1 --GCCGAAGUAGCCUUGGUUGGCGGAUGCACAGGAUGCUGGGGCGUUGGCAACGGCGAUUGCCAAGCCGAAGGAGAUCCCGGUGU---GGGCGUUGGCGA --(((((....((((..((((((....)).))....))..)))).)))))..((.((((.(((..((((..((....))))))..---.))))))).)). ( -38.90) >DroSim_CAF1 21518 95 - 1 --GCCGAUGUAGCCUUCGUGGGCGGAUGCACAGGAUGCUGGGGCGUUGGCAACGGCGAUUGCCAAGCCGAAGGAGAUCCCGGUGU---GGGCGUUGGCGA --((((((((.((((....))))....((((.((((.((..(((.(((((((......))))))))))...))..))))..))))---..)))))))).. ( -41.70) >DroEre_CAF1 20657 95 - 1 --GCUGAUGUAGCCUUGGUGGGAGGAUGCACGGGAUGCUGGGGCGUUGGCAACGGCGAUUGCCAAGCCGAAGGAGAUCCCGGUGU---GGGCGUUGGCGA --((..((((..((((.....))))((((.((((((.((..(((.(((((((......))))))))))...))..))))))))))---..))))..)).. ( -39.80) >DroYak_CAF1 22497 95 - 1 --GCCGACGUAGCCUUGGUGGGCGGAUGCACAGGAUGCUGGGGCGUUGGCAACGGCGAUUGCCAAGCCGAUGGAGAGCCCGGCGU---GGGCGUAGGCGA --(((.((((.((((....))))...........((((((((.(((((((...(((....)))..))))))).....))))))))---..)))).))).. ( -41.30) >consensus __GCCGAAGUAGCCUUGGUGGGCGGAUGCACAGGAUGCUGGGGCGUUGGCAACGGCGAUUGCCAAGCCGAAGGAGAUCCCGGUGU___GGGCGUUGGCGA ..(((......((((....)))).(((((.....((((((((((.(((((...(((....)))..)))))....).))))))))).....)))))))).. (-30.52 = -31.38 + 0.87)

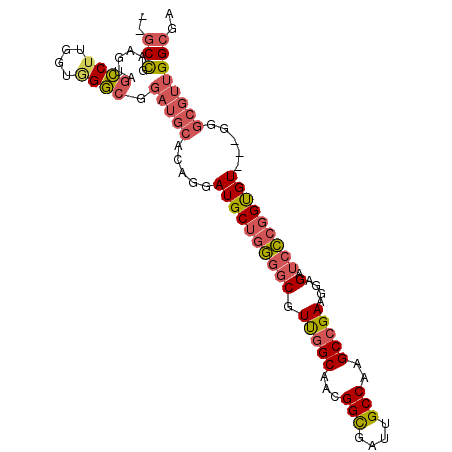
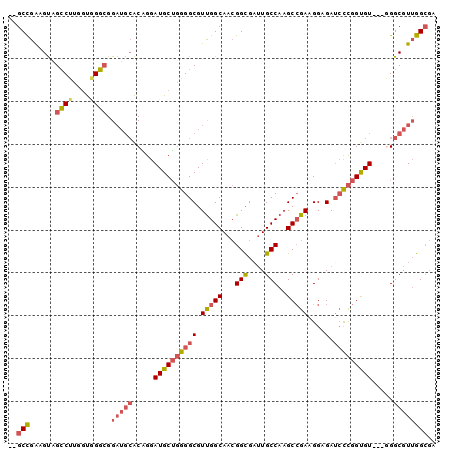
| Location | 10,773,611 – 10,773,710 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -21.10 |
| Energy contribution | -20.92 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10773611 99 + 22224390 UCCUUCGGCUUGGCAAUCACCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCCACUAAGGCUACUUCGGC---GGCCACGCCUACCCCGACACCAACGCC ......(((((((((((....)))))))).)))...((.....((((....(((......)))......(((---(....))))......).)))....)). ( -28.70) >DroVir_CAF1 27459 102 + 1 GCCAUCCGCGUGGCAGGCGCCUUUGCCGACGCCUGUGCAUCCAGCUGUGCCGGCUGUUAAGCCAGCUCCGGCAGCUGUUACGCCCGGCACGCCAGCUGGCAG (((....((((((((((((.(......).))))))).....(((((((((.((((....)))).))....))))))).)))))..)))..(((....))).. ( -45.90) >DroSec_CAF1 22991 99 + 1 UCCUUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCAACCAAGGCUACUUCGGC---GGCCACGCCUACCCCGACACCAACGCC ......(((((((((((....)))))))).)))...((.....((((....(((......)))......(((---(....))))......).)))....)). ( -29.20) >DroSim_CAF1 21541 99 + 1 UCCUUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCCACGAAGGCUACAUCGGC---GGCCACGCCUACCCCGACACCAACGCC .((((((((((((((((....)))))))).)))...((((...))))..........))))).......(((---(....)))).................. ( -31.30) >DroEre_CAF1 20680 99 + 1 UCCUUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCCGUGCAUCCUCCCACCAAGGCUACAUCAGC---GGCCACGCCUACGCCCACACCAUCGCC ......(((((((((((....)))))))).)))...((((...)))).............(((.......((---(....)))....)))............ ( -23.50) >DroYak_CAF1 22520 99 + 1 UCCAUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCCACCAAGGCUACGUCGGC---GGCCACGCCUACGCCGACACCAACGCC ......(((((((((((....)))))))).)))...((((...))))....(((......)))...((((((---(..........)))))))......... ( -35.10) >consensus UCCUUCGGCUUGGCAAUCGCCGUUGCCAACGCCCCAGCAUCCUGUGCAUCCGCCCACCAAGGCUACUUCGGC___GGCCACGCCUACCCCGACACCAACGCC ......(((((((((((....)))))))).)))............((....(((......)))....((((....(((...)))....)))).......)). (-21.10 = -20.92 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:45 2006