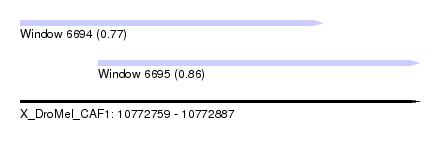
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,772,759 – 10,772,887 |
| Length | 128 |
| Max. P | 0.861219 |

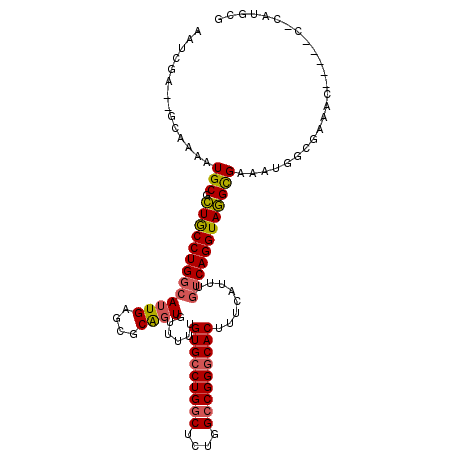
| Location | 10,772,759 – 10,772,856 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.77 |
| Mean single sequence MFE | -35.69 |
| Consensus MFE | -29.26 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10772759 97 + 22224390 ------------UGUACGCCAAGGCAAAGUGCUAAUAAAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUGCCUGACUCUGGCCGGGCACUUUCAUU ------------.....((....))((((((((..........--((((((.(((((((((.......)).)))))))))))))(((.......)))..)))))))).... ( -30.90) >DroSec_CAF1 22132 97 + 1 ------------UGUACGCCAAGGCAAAGUGCUAAUAAAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUUUUUUUGUGCCUGGCUCUGGCCGGGCACUUUCAUU ------------.....((((.((((..((((...........--.......))))))))))))..................(((((((((....)))))))))....... ( -35.37) >DroSim_CAF1 20682 97 + 1 ------------UGUACGCCAAGGCAAAGUGCUAAUAAAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUGCCUGGCUCUGGCCGGGCACUUUCAUU ------------.....((((.((((..((((...........--.......)))))))))))).........((((.....(((((((((....)))))))))...)))) ( -36.17) >DroEre_CAF1 19809 97 + 1 ------------AGUACGUCAAGGCAAAGUGCUAAUAAAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUGCCUGGCUCUCGCCGGGCACUUUCAUU ------------(((((...........)))))........((--(((...((((((...........)))))).)))))..(((((((((....)))))))))....... ( -33.80) >DroYak_CAF1 21672 97 + 1 ------------AGUACGCCAAGGCAAAGUGCUAAUAAAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUGUGUGCCUGGCUCUUGCCGGCCACUUUCGUU ------------.....(((.(((((..((((...........--.......)))))))))((((..((((.(((.((......))))))))).))))))).......... ( -32.97) >DroPer_CAF1 31655 111 + 1 AGUCCCAGCGAAAGUGUUCCAAGGCAAAGUGUUAAUAUACGGAGUGCGAAAUGCGUUACCUGGCAUUGAGUGCGCUCUUUGUGUGCCUGGCCCUCGCCGGGCACUGCCAAA .......((....)).......((((..((((....))))((((((((.(((((........)))))...))))))))....(((((((((....)))))))))))))... ( -44.90) >consensus ____________AGUACGCCAAGGCAAAGUGCUAAUAAAUCGA__GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUGCCUGGCUCUGGCCGGGCACUUUCAUU .................((((.(((..(((((....................))))))))))))..................(((((((((....)))))))))....... (-29.26 = -29.52 + 0.25)

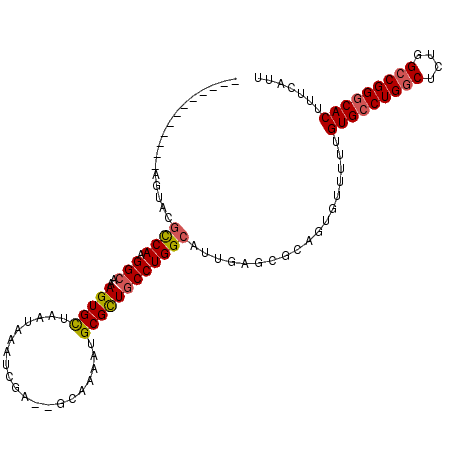

| Location | 10,772,784 – 10,772,887 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -32.92 |
| Energy contribution | -33.07 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10772784 103 + 22224390 AAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUGCCUGACUCUGGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGCUAAAC-----C-CAUGC- .....(--((....(((.((((((((((.((((...((((((..((.(((.......)))))))))))))))..))))))))))))).....)))....-----.-.....- ( -34.60) >DroSec_CAF1 22157 104 + 1 AAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUUUUUUUGUGCCUGGCUCUGGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGCGAAAC-----C-CAUGCG ......--((....(((.((((((((((.((((............(((((((((....))))))))).))))..)))))))))))))..((((......-----)-))))). ( -42.82) >DroSim_CAF1 20707 104 + 1 AAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUGCCUGGCUCUGGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGCGAAAC-----C-CAUGCG ......--((....(((.((((((((((.(((((.....))....(((((((((....)))))))))..)))..)))))))))))))..((((......-----)-))))). ( -43.00) >DroEre_CAF1 19834 110 + 1 AAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUGCCUGGCUCUCGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGGGAAUUUUGGGCGAGUUUG ..(((.--.((((((.((((((((((((.(((((.....))....(((((((((....)))))))))..)))..))))))))).))).........))))))..)))..... ( -42.50) >DroYak_CAF1 21697 105 + 1 AAUCGA--GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUGUGUGCCUGGCUCUUGCCGGCCACUUUCGUUUGUCAGGUAGGCGAAAUGGCGAAAAA----C-CAUUUG ......--......(((.((((((((((.(((((((((...))))))(((.(((....)))))).....)))..)))))))))))))((((((.......----)-))))). ( -37.80) >DroPer_CAF1 31692 106 + 1 UACGGAGUGCGAAAUGCGUUACCUGGCAUUGAGUGCGCUCUUUGUGUGCCUGGCCCUCGCCGGGCACUGCCAAAUCGCAGGUAAGUGGAUAGGGUCAGG-----C-CAGACG ...((((((((.(((((........)))))...))))))))..((.(((((((((((..(((.((.((((......))))))...)))..)))))))))-----.-)).)). ( -48.40) >consensus AAUCGA__GCAAAAUGCGCUGCCUGGCAUUGAGCGCAGUGUUUUUGUGCCUGGCUCUGGCCGGGCACUUUCAUUUGUCAGGUAGGCGAAAUGGCGAAAC_____C_CAUGCG ..............(((.(((((((((((((....))))......(((((((((....)))))))))........))))))))))))......................... (-32.92 = -33.07 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:42 2006