| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,768,741 – 10,768,883 |
| Length | 142 |
| Max. P | 0.994308 |

| Location | 10,768,741 – 10,768,845 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -19.31 |
| Energy contribution | -21.00 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10768741 104 + 22224390 UGCGAUUGUAUCAAAACAAGUUUUGAAUUAUCAAAAUUGGUUCCAUUUAUUUUUAUACAACCUUGACCUAUUUUCAAGGACCAAUAAGAUUGGACCCCACAUUA ...(((.((.((((((....)))))))).)))......((.((((.....((((((....((((((.......))))))....)))))).)))).))....... ( -19.50) >DroSec_CAF1 18204 104 + 1 UGCGAUUGUAUAAAAACAUGUUUUGAAUUAUCAAAAUAUGUUCCAUUUAUUUUUAUACAACCUUGACCUAUUUUCAAGGACCAAUAAGAUUGGACCCCACAUAA .....(((((((((((((((((((((....))))))))))).........))))))))))((((((.......)))))).(((((...)))))........... ( -25.30) >DroSim_CAF1 16386 103 + 1 UGCGAUUGUA-AAAAACAAGUUUUGAAUUAUCAAAAUAUGUUCCAUUUAUUUUUAUACAACCUUGACCUAUUUUCAAGGACCAAUAAGAUUGGACCCCACAUAA ...((..(((-((.((((.(((((((....))))))).))))...)))))..))......((((((.......)))))).(((((...)))))........... ( -17.70) >DroYak_CAF1 17870 100 + 1 UGCGAUUGUAUAAAAACAAGUC-CGAAAUUUCAAAAUAUGUUUGAUUUAUUUUUAUGCAACCUUGACCUAUUUUCAAGGACCAAUAAUAUUGGACCCCGCA--- .(((.(((((((((((.(((((-.((.((........)).)).))))).)))))))))))((((((.......)))))).(((((...)))))....))).--- ( -24.60) >consensus UGCGAUUGUAUAAAAACAAGUUUUGAAUUAUCAAAAUAUGUUCCAUUUAUUUUUAUACAACCUUGACCUAUUUUCAAGGACCAAUAAGAUUGGACCCCACAUAA .....(((((((((((((((((((((....))))))))))).........))))))))))((((((.......)))))).(((((...)))))........... (-19.31 = -21.00 + 1.69)

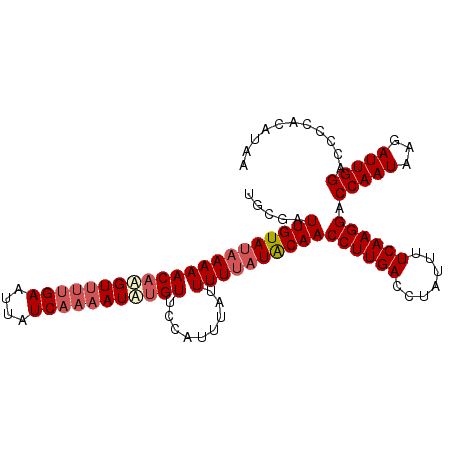

| Location | 10,768,741 – 10,768,845 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -21.64 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10768741 104 - 22224390 UAAUGUGGGGUCCAAUCUUAUUGGUCCUUGAAAAUAGGUCAAGGUUGUAUAAAAAUAAAUGGAACCAAUUUUGAUAAUUCAAAACUUGUUUUGAUACAAUCGCA ...(((((((.(((((...))))).)))).............((((((((.(((((((.((....)).((((((....)))))).))))))).))))))))))) ( -24.00) >DroSec_CAF1 18204 104 - 1 UUAUGUGGGGUCCAAUCUUAUUGGUCCUUGAAAAUAGGUCAAGGUUGUAUAAAAAUAAAUGGAACAUAUUUUGAUAAUUCAAAACAUGUUUUUAUACAAUCGCA ((((.(((((.(((((...))))).)))))...)))).....((((((((((.........((((((.((((((....)))))).))))))))))))))))... ( -26.30) >DroSim_CAF1 16386 103 - 1 UUAUGUGGGGUCCAAUCUUAUUGGUCCUUGAAAAUAGGUCAAGGUUGUAUAAAAAUAAAUGGAACAUAUUUUGAUAAUUCAAAACUUGUUUUU-UACAAUCGCA ((((.(((((.(((((...))))).)))))...)))).....(((((((...........((((((..((((((....))))))..)))))).-)))))))... ( -22.80) >DroYak_CAF1 17870 100 - 1 ---UGCGGGGUCCAAUAUUAUUGGUCCUUGAAAAUAGGUCAAGGUUGCAUAAAAAUAAAUCAAACAUAUUUUGAAAUUUCG-GACUUGUUUUUAUACAAUCGCA ---.((((((.(((((...))))).)))((((((((((((.(((((.....((((((.........))))))..)))))..-))))))))))))......))). ( -23.70) >consensus UUAUGUGGGGUCCAAUCUUAUUGGUCCUUGAAAAUAGGUCAAGGUUGUAUAAAAAUAAAUGGAACAUAUUUUGAUAAUUCAAAACUUGUUUUUAUACAAUCGCA ...(((((((.(((((...))))).)))).............((((((((((((((((..........((((((....)))))).))))))))))))))))))) (-21.64 = -22.32 + 0.69)


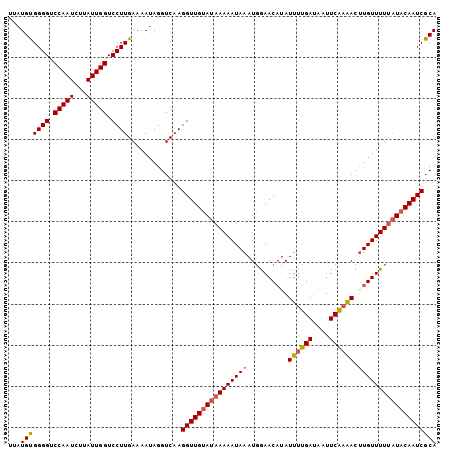
| Location | 10,768,777 – 10,768,883 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -15.01 |
| Consensus MFE | -9.90 |
| Energy contribution | -9.52 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10768777 106 + 22224390 UUGGUUCCAUUUAUUUUUAUACAACCUUGACCUAUUUUCAAGGACCAAUAAGAUUGGACCCCACAUUAACUUAGAAAACAAUACUUGCCAUGUUCAAUUUUAUUCC- .((((......(((....)))...((((((.......))))))))))((((((((((((....((....................))....))))))))))))...- ( -15.85) >DroSec_CAF1 18240 95 + 1 UAUGUUCCAUUUAUUUUUAUACAACCUUGACCUAUUUUCAAGGACCAAUAAGAUUGGACCCCACAUAAACACAGAAAACAACAAUU------------CGUAUUCCA (((((((((.....((((((....((((((.......))))))....)))))).))))....)))))......(((.((.......------------.)).))).. ( -14.20) >DroSim_CAF1 16421 95 + 1 UAUGUUCCAUUUAUUUUUAUACAACCUUGACCUAUUUUCAAGGACCAAUAAGAUUGGACCCCACAUAAACACAGAAAGCAACAAUU------------CGUAUUCCA (((((((((.....((((((....((((((.......))))))....)))))).))))....)))))......(((.((.......------------.)).))).. ( -14.50) >DroYak_CAF1 17905 89 + 1 UAUGUUUGAUUUAUUUUUAUGCAACCUUGACCUAUUUUCAAGGACCAAUAAUAUUGGACCCCGCA------CAGGAAACAACAAUU------------CUCAUUUUA ..((((.............(((..((((((.......)))))).(((((...))))).....)))------..(....)))))...------------......... ( -15.50) >consensus UAUGUUCCAUUUAUUUUUAUACAACCUUGACCUAUUUUCAAGGACCAAUAAGAUUGGACCCCACAUAAACACAGAAAACAACAAUU____________CGUAUUCCA .............(((((......((((((.......)))))).(((((...)))))................)))))............................. ( -9.90 = -9.52 + -0.37)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:40 2006