
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,766,095 – 10,766,231 |
| Length | 136 |
| Max. P | 0.999921 |

| Location | 10,766,095 – 10,766,191 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -29.76 |
| Energy contribution | -30.20 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10766095 96 + 22224390 ACAUCUUAAGUUGGUGGUAAACGAUGCAGCGGCAUCAAAAUAAUUUUGAUACUAUGUGCAUCAUGGAAAC-GAAACAGCGGUUGCACCGUUUACAAC .((((.......))))(((((((.((((((.((((((((.....)))))).(((((.....)))))....-......)).)))))).)))))))... ( -32.60) >DroSec_CAF1 15477 96 + 1 ACAUCUUAUGUUGGUGGUAAACGAUGCAGCGGCAUCAAAAUAAUUUUGAUACUAUGUGAAUCAUGGAAAC-GAAACUGCGGUUGCACCGUUUACAAC .((((.......))))(((((((.((((((.((((((((.....)))))).(((((.....)))))....-......)).)))))).)))))))... ( -32.40) >DroSim_CAF1 13662 96 + 1 ACAUCUUAAGUUGGUGGUAAACGAUGCAGCGGCAUCAAAAUAAUUUUGAUACUAUGUGCAUCAUGGAAAC-GAAACUGCGGUUGCACCGUUUACAAC .((((.......))))(((((((.((((((.((((((((.....)))))).(((((.....)))))....-......)).)))))).)))))))... ( -32.40) >DroEre_CAF1 13394 96 + 1 ACAUCCAAAGUUGGUG-UAAACGAUGCAGCGGCAUCAAAACAAUUUUGAUACUAUGUGCAUCAUGGAAACUGAAACGGCGGCUGCAUCGUUUACAAC ....(((....)))((-(((((((((((((.((((((((.....)))))).(((((.....)))))...........)).))))))))))))))).. ( -38.20) >DroYak_CAF1 15199 97 + 1 ACAUCCAAAGUUGGUGGUAAACGAUGCAGCGGCAGCAAAAUAAUUUUGAUACUAUGUGCAGCAUGGAAACUGAAACGGCGGUCGCAUCGUUUACAAC .((.(((....)))))(((((((((((.((.((..((((.....))))...(((((.....)))))...........)).)).)))))))))))... ( -30.70) >consensus ACAUCUUAAGUUGGUGGUAAACGAUGCAGCGGCAUCAAAAUAAUUUUGAUACUAUGUGCAUCAUGGAAAC_GAAACGGCGGUUGCACCGUUUACAAC .((((.......))))(((((((.((((((.((((((((.....)))))).(((((.....)))))...........)).)))))).)))))))... (-29.76 = -30.20 + 0.44)



| Location | 10,766,095 – 10,766,191 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10766095 96 - 22224390 GUUGUAAACGGUGCAACCGCUGUUUC-GUUUCCAUGAUGCACAUAGUAUCAAAAUUAUUUUGAUGCCGCUGCAUCGUUUACCACCAACUUAAGAUGU ((.((((((((((((...(((((.((-((....)))).)))....((((((((.....)))))))).)))))))))))))).))............. ( -28.20) >DroSec_CAF1 15477 96 - 1 GUUGUAAACGGUGCAACCGCAGUUUC-GUUUCCAUGAUUCACAUAGUAUCAAAAUUAUUUUGAUGCCGCUGCAUCGUUUACCACCAACAUAAGAUGU ((.((((((((((((...((.(..((-((....))))..).....((((((((.....)))))))).)))))))))))))).))............. ( -27.60) >DroSim_CAF1 13662 96 - 1 GUUGUAAACGGUGCAACCGCAGUUUC-GUUUCCAUGAUGCACAUAGUAUCAAAAUUAUUUUGAUGCCGCUGCAUCGUUUACCACCAACUUAAGAUGU ((.((((((((((((...((.((.((-((....)))).)).....((((((((.....)))))))).)))))))))))))).))............. ( -28.40) >DroEre_CAF1 13394 96 - 1 GUUGUAAACGAUGCAGCCGCCGUUUCAGUUUCCAUGAUGCACAUAGUAUCAAAAUUGUUUUGAUGCCGCUGCAUCGUUUA-CACCAACUUUGGAUGU ..(((((((((((((((.((.(((.((((((...((((((.....))))))))))))....))))).)))))))))))))-))(((....))).... ( -37.90) >DroYak_CAF1 15199 97 - 1 GUUGUAAACGAUGCGACCGCCGUUUCAGUUUCCAUGCUGCACAUAGUAUCAAAAUUAUUUUGCUGCCGCUGCAUCGUUUACCACCAACUUUGGAUGU ...((((((((((((.(.((.((...(((((..((((((....))))))..))))).....)).)).).))))))))))))..(((....))).... ( -26.50) >consensus GUUGUAAACGGUGCAACCGCAGUUUC_GUUUCCAUGAUGCACAUAGUAUCAAAAUUAUUUUGAUGCCGCUGCAUCGUUUACCACCAACUUAAGAUGU ((.((((((((((((...((.............(((.....))).((((((((.....)))))))).)))))))))))))).))............. (-23.58 = -23.58 + 0.00)

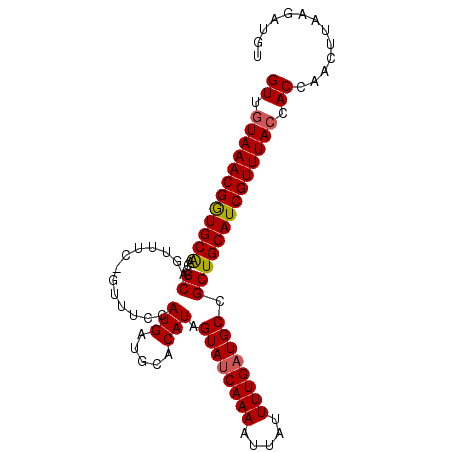

| Location | 10,766,126 – 10,766,231 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -27.84 |
| Energy contribution | -27.56 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10766126 105 - 22224390 AACGUGGAGUGAUUCACUUUGGAUCUCGUUGUUGAAGCUGGUUGUAAACGGUGCAACCGCUGUUUC-GUUUCCAUGAUGCACAUAGUAUCAAAAUUAUUUUGAUGC ..(((((((.(((((.....))))).)......(((((((((((((.....))))))))..)))))-...)))))).........((((((((.....)))))))) ( -31.50) >DroSec_CAF1 15508 105 - 1 AACGUGGAGUGAUUCACUUUGGAUCUCGUUGAUGGAGCUGGUUGUAAACGGUGCAACCGCAGUUUC-GUUUCCAUGAUUCACAUAGUAUCAAAAUUAUUUUGAUGC ...(((((....)))))..((((((..(..((((((((((((((((.....)))))))..))))))-)))..)..))))))....((((((((.....)))))))) ( -34.70) >DroSim_CAF1 13693 105 - 1 AACGUGGAGUGAUUCACAUUGGAUCUCGUUGAUGGAGCUGGUUGUAAACGGUGCAACCGCAGUUUC-GUUUCCAUGAUGCACAUAGUAUCAAAAUUAUUUUGAUGC ..(((((((.(((((.....))))).)...((((((((((((((((.....)))))))..))))))-))))))))).........((((((((.....)))))))) ( -33.90) >DroEre_CAF1 13424 106 - 1 AACGUGAAGUGACUCACUUUGGAUUUCGUUGGUGAAGCCGGUUGUAAACGAUGCAGCCGCCGUUUCAGUUUCCAUGAUGCACAUAGUAUCAAAAUUGUUUUGAUGC ........(((..(((...((((.......(.((((((((((((((.....))))))))..)))))).).)))))))..)))...((((((((.....)))))))) ( -28.60) >DroYak_CAF1 15230 106 - 1 AACGUGGAGUGAUUCACUUUGGAUCUCGUUGGUGAAGCUGGUUGUAAACGAUGCGACCGCCGUUUCAGUUUCCAUGCUGCACAUAGUAUCAAAAUUAUUUUGCUGC ..(((((((.(((((.....))))).).....((((((((((((((.....))))))))..))))))...))))))..(((((.((((.......)))).)).))) ( -28.60) >consensus AACGUGGAGUGAUUCACUUUGGAUCUCGUUGAUGAAGCUGGUUGUAAACGGUGCAACCGCAGUUUC_GUUUCCAUGAUGCACAUAGUAUCAAAAUUAUUUUGAUGC ..(((((((.(((((.....))))).)......(((((((((((((.....))))))))..)))))....)))))).........((((((((.....)))))))) (-27.84 = -27.56 + -0.28)

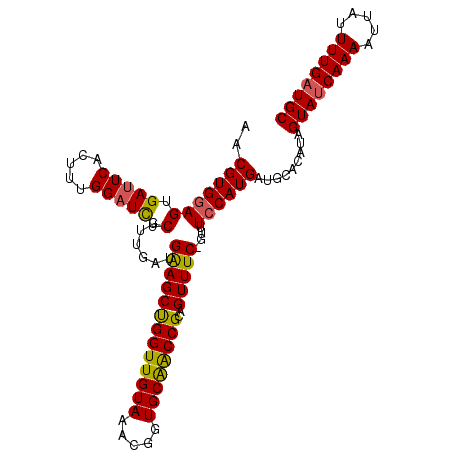
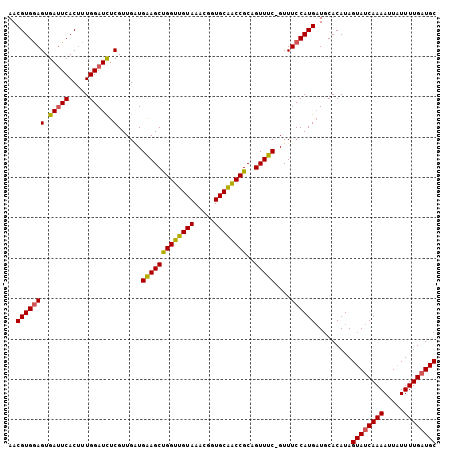
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:37 2006