| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,765,511 – 10,765,617 |
| Length | 106 |
| Max. P | 0.849991 |

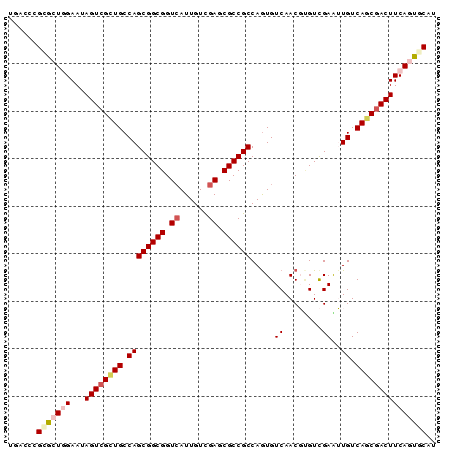
| Location | 10,765,511 – 10,765,601 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -30.90 |
| Energy contribution | -31.98 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10765511 90 + 22224390 UGACCCGCGCUCGAAUAGUCGAUGCCAGCGGCGGUCAUUGUCGAGCGCCGCCAGUGUCAACUUGUCGAAAUGUCAGCGACUUCAGUGCAU ......(((((.(((.(((.(((((..((((((.((......)).))))))..))))).))).((((.........)))))))))))).. ( -34.00) >DroGri_CAF1 14202 90 + 1 UGACCCGGGCUGGAAUAGUCGCUGCCAGCGGCGGUCAUUGUCGAGCGCCGCCAGUGUCAACGCGUCGAGUUGUCAGCGACUUCAGUUCAU ......(((((((...((((((((.((((((((.((......)).))))))..(((....))).......)).))))))))))))))).. ( -39.80) >DroWil_CAF1 9905 90 + 1 UGACCCGCAAUGGAAUAGUCGUUGCCAGCGGCGGUCAUUGUUGAGCGCCGCCAGUGUCAACGUGUCGAAUUGUCAGCAACUUCAGUGCAU ((((.((((((((.....)))))))..((((((.(((....))).))))))..).))))..(..(.((((((....))).))).)..).. ( -31.80) >DroYak_CAF1 14635 90 + 1 UGACCCGCGCUCGAAUAGUCGCUGCCAGCGGCGGUCAUUGUCGAGCGCCGCCAGUGUCAACUAGUUGAAAUGUCAGCGACUUCAGUGCAU ......(((((.((..((((((((.((((((((.((......)).)))))).....((((....))))..)).))))))))))))))).. ( -39.90) >DroMoj_CAF1 14422 90 + 1 UGACCCGGGCUGGAAUAGUCGCUGCCAGCGGCGGUGAUUGUUGAGCGCCGCCAAUGUCAACGCGUCGAGUUGUCAGCGACUUCAGUGCAC ......(.(((((...((((((((.(((((((((((.........))))))).((((....))))...)))).))))))))))))).).. ( -38.70) >DroAna_CAF1 23406 90 + 1 UGACCCGCGAUGGAAUAGUCGCUGCCAGCGGCGGUCAUUGUCGAGCGCCGCCAGUGUCAACGUGUCGAGUUGUCAGCGACUUCAGUCCAU .........(((((..(((((((((..((((((.((......)).))))))..))(.((((.......)))).))))))))....))))) ( -36.80) >consensus UGACCCGCGCUGGAAUAGUCGCUGCCAGCGGCGGUCAUUGUCGAGCGCCGCCAGUGUCAACGUGUCGAAUUGUCAGCGACUUCAGUGCAU ......(((((((...((((((((.((((((((.((......)).)))))).....((........))..)).))))))))))))))).. (-30.90 = -31.98 + 1.09)


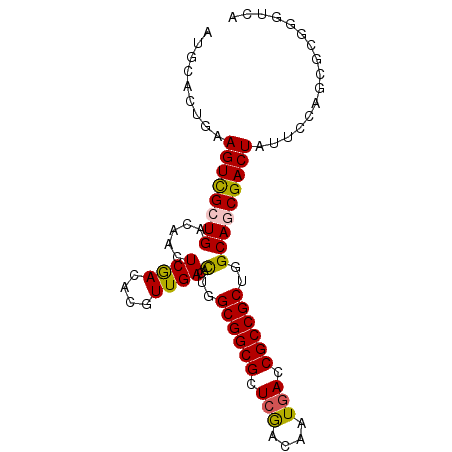
| Location | 10,765,511 – 10,765,601 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -29.39 |
| Energy contribution | -29.25 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10765511 90 - 22224390 AUGCACUGAAGUCGCUGACAUUUCGACAAGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAUCGACUAUUCGAGCGCGGGUCA .(((.(((((((((.........)))).((((((..(..((((((.((......)).))))))..)..)))))).))).)).)))..... ( -32.90) >DroGri_CAF1 14202 90 - 1 AUGAACUGAAGUCGCUGACAACUCGACGCGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAGCGACUAUUCCAGCCCGGGUCA .(((.....((((((((.((((.......))))...(..((((((.((......)).))))))..)))))))))..(((.....)))))) ( -34.40) >DroWil_CAF1 9905 90 - 1 AUGCACUGAAGUUGCUGACAAUUCGACACGUUGACACUGGCGGCGCUCAACAAUGACCGCCGCUGGCAACGACUAUUCCAUUGCGGGUCA .((((.((..((((.........)))).(((((...(..((((((.(((....))).))))))..)))))).......)).))))..... ( -28.80) >DroYak_CAF1 14635 90 - 1 AUGCACUGAAGUCGCUGACAUUUCAACUAGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAGCGACUAUUCGAGCGCGGGUCA .(((.((((((((((((.....((((....))))..(..((((((.((......)).))))))..)))))))))..)).)).)))..... ( -36.70) >DroMoj_CAF1 14422 90 - 1 GUGCACUGAAGUCGCUGACAACUCGACGCGUUGACAUUGGCGGCGCUCAACAAUCACCGCCGCUGGCAGCGACUAUUCCAGCCCGGGUCA .........((((((((.((((.......))))...(..((((((............))))))..)))))))))..(((.....)))... ( -29.90) >DroAna_CAF1 23406 90 - 1 AUGGACUGAAGUCGCUGACAACUCGACACGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAGCGACUAUUCCAUCGCGGGUCA (((((....((((((((.((((.......))))...(..((((((.((......)).))))))..)))))))))..)))))......... ( -36.80) >consensus AUGCACUGAAGUCGCUGACAACUCGACACGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAGCGACUAUUCCAGCGCGGGUCA .........((((((((.....((((....))))..(..((((((.(((....))).))))))..)))))))))................ (-29.39 = -29.25 + -0.14)


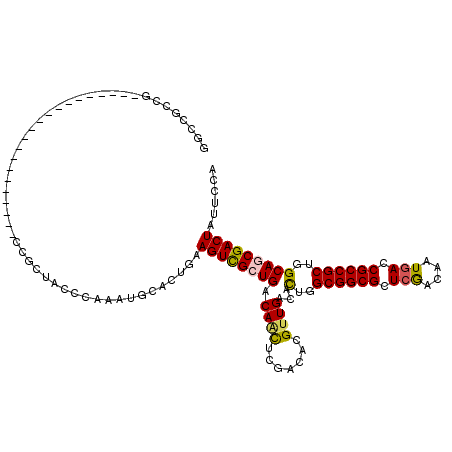
| Location | 10,765,521 – 10,765,617 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -29.51 |
| Energy contribution | -29.35 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10765521 96 - 22224390 UGCCACCG------------------------CUACCCAAAUGCACUGAAGUCGCUGACAUUUCGACAAGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAUCGACUAUUCGA ........------------------------......(((((((.((....)).)).)))))(((..((((((..(..((((((.((......)).))))))..)..))))))..))). ( -29.60) >DroPse_CAF1 30109 108 - 1 CGCCGCCG--------UUCA----CUCCGCCGCGACCCAAAUGCACUGAAGUCGCUGACAGCUCGACGCGUUGACACUGGCGGCGCUCGACGAUGACCGCCGCUGGCAGCGACUAUUCAA (((.((.(--------....----..).)).)))............(((((((((((.((((.......))))...(..((((((.((......)).))))))..)))))))))..))). ( -39.40) >DroGri_CAF1 14212 99 - 1 UGGCGCCA---------------------CCGCCGCCCAAAUGAACUGAAGUCGCUGACAACUCGACGCGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAGCGACUAUUCCA .((((...---------------------.))))...............((((((((.((((.......))))...(..((((((.((......)).))))))..)))))))))...... ( -39.10) >DroWil_CAF1 9915 120 - 1 GGCCGCCUGCACCACCUGCACCACCACCUCCGCUACCCAAAUGCACUGAAGUUGCUGACAAUUCGACACGUUGACACUGGCGGCGCUCAACAAUGACCGCCGCUGGCAACGACUAUUCCA (((.((.((((.....))))...........((.........))......)).)))............(((((...(..((((((.(((....))).))))))..))))))......... ( -30.00) >DroMoj_CAF1 14432 99 - 1 GGCCAGCA---------------------CCACCGCCCAAGUGCACUGAAGUCGCUGACAACUCGACGCGUUGACAUUGGCGGCGCUCAACAAUCACCGCCGCUGGCAGCGACUAUUCCA ((...(((---------------------(..........)))).....((((((((.((((.......))))...(..((((((............))))))..)))))))))...)). ( -33.90) >DroAna_CAF1 23416 108 - 1 GUCCCCCG--------CCCA----GUCCUCCACUCCCCAGAUGGACUGAAGUCGCUGACAACUCGACACGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAGCGACUAUUCCA ........--------..((----((((..(........)..)))))).((((((((.((((.......))))...(..((((((.((......)).))))))..)))))))))...... ( -42.30) >consensus GGCCGCCG_____________________CCGCUACCCAAAUGCACUGAAGUCGCUGACAACUCGACACGUUGACACUGGCGGCGCUCGACAAUGACCGCCGCUGGCAGCGACUAUUCCA .................................................((((((((.((((.......))))...(..((((((.(((....))).))))))..)))))))))...... (-29.51 = -29.35 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:35 2006