
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,762,490 – 10,762,674 |
| Length | 184 |
| Max. P | 0.977280 |

| Location | 10,762,490 – 10,762,598 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -44.73 |
| Consensus MFE | -36.15 |
| Energy contribution | -37.23 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10762490 108 + 22224390 UCCUACUACACAUUGGCCGCUGCAGCUGCGGCUCAUCCGCAUGCCCAUGGCCACGCCCACACGCAUGCGCAUGCCGCCCACUAUGCCGCCGCCGCUGCGGCAGCUGAC ............(..((.((((((((.(((((......((((((....(((...))).....))))))(((((........))))).))))).)))))))).))..). ( -49.20) >DroSec_CAF1 12181 108 + 1 UCCUACUACACAUUGGCCGCUGCAGCUGCUGCUCAUCCGCAUGCCCAUGGCCACGCCCACACGCAUGCGCAUGCCGCCCACUAUGCCGCCGCCGCUGCGACGGCUGAC ............(..((((.((((((.((.((......((((((....(((...))).....))))))(((((........))))).)).)).)))))).))))..). ( -42.60) >DroSim_CAF1 9748 108 + 1 UCCUACUACACAUUGGCCGCUGCAGCUGCUGCUCAUCCGCAUGCCCAUGGCCACGCCCACACGCAUGCGCAUGCCGCCCACUAUGCCGCCGCCGCUGCGGCGGCUGAC ............(..(((((((((((.((.((......((((((....(((...))).....))))))(((((........))))).)).)).)))))))))))..). ( -49.20) >DroEre_CAF1 10688 108 + 1 UCCUACUACACAUUGGCCGCUGCAGCUGCCGCUCAUCCGCAUGCCCAUGGCCACGCCCACACGCAUGCGCAUGCCGCCCACUACGCCGCCGCCGCUGCGGCAGCCGAC ............(((((.((((((((.((.((.(((.(((((((....(((...))).....))))))).)))..((.......)).)).)).)))))))).))))). ( -44.80) >DroYak_CAF1 12839 108 + 1 UCCUACUACACAUUGGCCGCUGCAGCUGCCGCUCAUCCGCAUGCUCAUGGCCACGCCCACACGCAUGCGCAUGCCGCCCACUAUGCCGCCGCCGCUGCGGCAGCUGAC ............(..((.((((((((.((.((......((((((....(((...))).....))))))(((((........))))).)).)).)))))))).))..). ( -43.10) >DroAna_CAF1 20504 108 + 1 UCGUACUACACCCUGGCGGCCGCUGCUGCCGGUCAUCCGCAUGCCCAUGGCCACGCCCACGCCCAUGCCCAUGCCGCCCACUAUGCUGCCGCCGCCGCGUCAGCGGAC ............((((((((....))))))))...((((((((..(((((.(........).)))))..)))))..........((((.(((....))).))))))). ( -39.50) >consensus UCCUACUACACAUUGGCCGCUGCAGCUGCCGCUCAUCCGCAUGCCCAUGGCCACGCCCACACGCAUGCGCAUGCCGCCCACUAUGCCGCCGCCGCUGCGGCAGCUGAC ............(((((.((((((((.((.((.(((.(((((((....(((...))).....))))))).)))..((.......)).)).)).)))))))).))))). (-36.15 = -37.23 + 1.08)

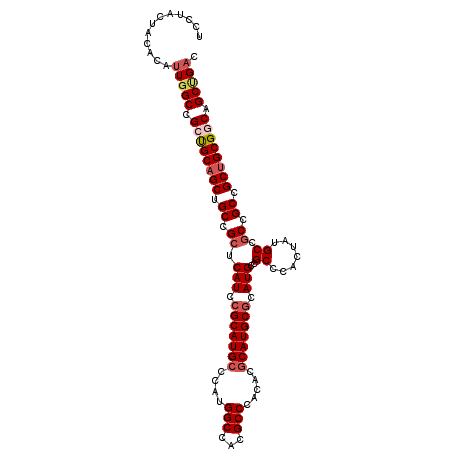

| Location | 10,762,490 – 10,762,598 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -52.72 |
| Consensus MFE | -47.11 |
| Energy contribution | -47.17 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10762490 108 - 22224390 GUCAGCUGCCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAUGCGGAUGAGCCGCAGCUGCAGCGGCCAAUGUGUAGUAGGA ....(((((.(((((.(((((.((.......))..(((.(((((((.(((((......))))).))))))).))).))))).))))).)))))............... ( -55.40) >DroSec_CAF1 12181 108 - 1 GUCAGCCGUCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAUGCGGAUGAGCAGCAGCUGCAGCGGCCAAUGUGUAGUAGGA ....((((((((....))))))))(((.(((.((.(((.(((((((.(((((......))))).))))))).))).((((...))))..)).))).)))......... ( -51.90) >DroSim_CAF1 9748 108 - 1 GUCAGCCGCCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAUGCGGAUGAGCAGCAGCUGCAGCGGCCAAUGUGUAGUAGGA ....((((((((....))))))))(((.(((.((.(((.(((((((.(((((......))))).))))))).))).((((...))))..)).))).)))......... ( -54.60) >DroEre_CAF1 10688 108 - 1 GUCGGCUGCCGCAGCGGCGGCGGCGUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAUGCGGAUGAGCGGCAGCUGCAGCGGCCAAUGUGUAGUAGGA ...((((((.(((((.((.((..(((.....))).(((.(((((((.(((((......))))).))))))).))).)).)).))))).)))))).............. ( -56.20) >DroYak_CAF1 12839 108 - 1 GUCAGCUGCCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGAGCAUGCGGAUGAGCGGCAGCUGCAGCGGCCAAUGUGUAGUAGGA (((.((((((((....)))))))).......((..(((.(((((((..((((......))))..))))))).))).))))).((((((.((.....)))))))).... ( -50.90) >DroAna_CAF1 20504 108 - 1 GUCCGCUGACGCGGCGGCGGCAGCAUAGUGGGCGGCAUGGGCAUGGGCGUGGGCGUGGCCAUGGGCAUGCGGAUGACCGGCAGCAGCGGCCGCCAGGGUGUAGUACGA (.((((....)))))((((((.((...((.(.((((((..(((((..(((((......)))))..)))))..))).))).).)).)).)))))).............. ( -47.30) >consensus GUCAGCUGCCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAUGCGGAUGAGCGGCAGCUGCAGCGGCCAAUGUGUAGUAGGA ....((((((((....))))))))......(((..(((.(((((((.(((((......))))).))))))).))).((.((....)).)).))).............. (-47.11 = -47.17 + 0.06)

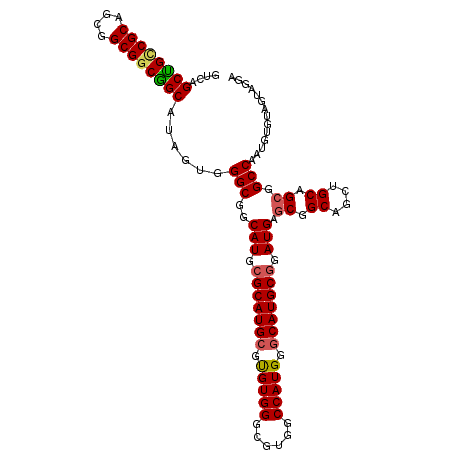

| Location | 10,762,530 – 10,762,638 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 91.91 |
| Mean single sequence MFE | -53.10 |
| Consensus MFE | -47.01 |
| Energy contribution | -47.07 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10762530 108 - 22224390 CAGCGGCUGCAGCCGCUCCCAGUAGCUGUCCCAUAUUGAAGUCAGCUGCCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAU .((((((....))))))((((...((((.(((((..((....))((((((((....))))))))...)))))))))..((.(((((......)))))))..))))... ( -54.60) >DroSec_CAF1 12221 108 - 1 CAGCGGCUGCAGCCGCACCCAGUAGCUGUCCCAUAUUAAAGUCAGCCGUCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAU ..(((((....))))).((((...((..((((((((........((((((((....))))))))...((..(((.....)))..))))))))).)..))..))))... ( -51.50) >DroSim_CAF1 9788 108 - 1 CAGCGGCUGCAGCCGCACCCAGUAGCUGUCCCAUAUUAAAGUCAGCCGCCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAU ..(((((....))))).((((...((..((((((((........((((((((....))))))))...((..(((.....)))..))))))))).)..))..))))... ( -54.20) >DroEre_CAF1 10728 108 - 1 CAGCGGCUGCAGCCGCUCCCAGUAGCUGCCCCAUAUUGAAGUCGGCUGCCGCAGCGGCGGCGGCGUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAU .((((((....))))))((((...((..((((((((........((((((((....))))))))(((.((.((.....)).)))))))))))).)..))..))))... ( -55.40) >DroYak_CAF1 12879 108 - 1 CAGCGGCUGCAGCCGCUCCCAGUAGCUGCCCCAUAUUGAAGUCAGCUGCCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGAGCAU .....(((.(((((((.(((....((((((((....((....))((((((((....))))))))...).)))))))(((((....)))))))).)))))..))))).. ( -54.00) >DroAna_CAF1 20544 108 - 1 CUGCCGCAGCUGCUGCCCCCAAAAGCUGGCUCAUAUUGAAGUCCGCUGACGCGGCGGCGGCAGCAUAGUGGGCGGCAUGGGCAUGGGCGUGGGCGUGGCCAUGGGCAU (((((.((((((((((((((...(((.(((((.....).)))).)))...).)).)))))))))....)))))))(((((.((((........)))).)))))..... ( -48.90) >consensus CAGCGGCUGCAGCCGCUCCCAGUAGCUGUCCCAUAUUGAAGUCAGCUGCCGCAGCGGCGGCGGCAUAGUGGGCGGCAUGCGCAUGCGUGUGGGCGUGGCCAUGGGCAU .....(((...(((((.(((....((((.(((((..((....))((((((((....))))))))...)))))))))(((((....)))))))).)))))....))).. (-47.01 = -47.07 + 0.06)
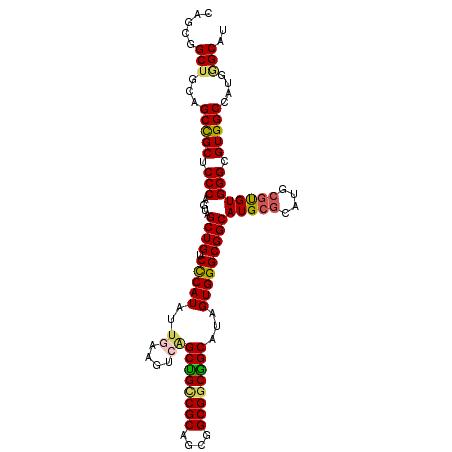
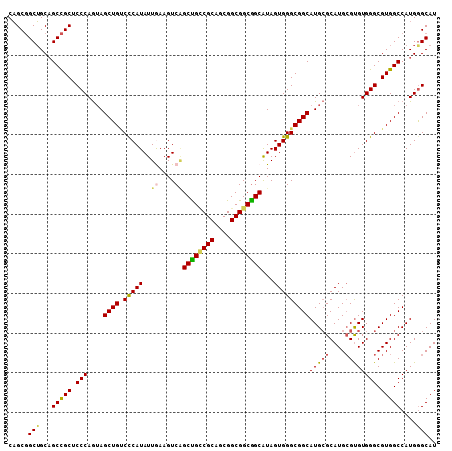
| Location | 10,762,569 – 10,762,674 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -51.73 |
| Consensus MFE | -34.32 |
| Energy contribution | -35.93 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10762569 105 + 22224390 ACUAUGCCGCCGCCGCUGCGGCAGCUGACUUCAAUAUGGGACAGCUACUGGGAGCGGCUGCAGCCGCUGCCGCCUCUGGAGAUCCCGCUGCCCAGCAUGCGGCUG ........(((((.((((.((((((((....))....((((...((.(..(..(((((.((....)).)))))..)..))).))))))))))))))..))))).. ( -55.30) >DroSec_CAF1 12260 105 + 1 ACUAUGCCGCCGCCGCUGCGACGGCUGACUUUAAUAUGGGACAGCUACUGGGUGCGGCUGCAGCCGCUGCCGCCUCUGGAGAUCCCGCUGCCCAGCAUGCGGCUG .....(((((.((((......))))...........((((.((((....(((.(((((.((....)).)))))))).((.....))))))))))....))))).. ( -46.20) >DroEre_CAF1 10767 105 + 1 ACUACGCCGCCGCCGCUGCGGCAGCCGACUUCAAUAUGGGGCAGCUACUGGGAGCGGCUGCAGCCGCUGCCGCCUCUGGAGAUCCGGCUGCACAGCAUGCGGCUG ........(((((.((((..(((((((.(((((....(((((((...))((.((((((....)))))).))))))))))))...))))))).))))..))))).. ( -56.00) >DroWil_CAF1 7617 102 + 1 AUUAUGCAGCCGCUGCUGCGGCAGCUGACUUCAAUAUGGGACAAUUGCUGGGAGCUGCGGCAGCUGCUGCUGCCACUGGCGACGCAUCGUCGAUGC---CAGCUG .....(((((.((.(((((.((((((..(..((((........))))..)..)))))).))))).)).)))))..(((((((((...))))...))---)))... ( -49.00) >DroYak_CAF1 12918 105 + 1 ACUAUGCCGCCGCCGCUGCGGCAGCUGACUUCAAUAUGGGGCAGCUACUGGGAGCGGCUGCAGCCGCUGCCGCUUCUGGAGAUCCGGCUGCACAGCAUGCGGCUG .....(((((.((.(.(((((((((((.((((.....))))))))).(..((((((((.((....)).))))))))..).......))))))).))..))))).. ( -54.60) >DroAna_CAF1 20583 105 + 1 ACUAUGCUGCCGCCGCCGCGUCAGCGGACUUCAAUAUGAGCCAGCUUUUGGGGGCAGCAGCUGCGGCAGCCGCCUCCGGGGAUCCGGCGGCCCAACAUGCCGCCG ....(((((((.((.((((....))))..........(((....)))..)).)))))))...((((((...(((.((((....)))).)))......)))))).. ( -49.30) >consensus ACUAUGCCGCCGCCGCUGCGGCAGCUGACUUCAAUAUGGGACAGCUACUGGGAGCGGCUGCAGCCGCUGCCGCCUCUGGAGAUCCGGCUGCCCAGCAUGCGGCUG ........((.((((((.(((.(((((.(((......))).))))).))).).))))).))(((((((((.((..((((....))))..))...))).)))))). (-34.32 = -35.93 + 1.61)



| Location | 10,762,569 – 10,762,674 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -52.87 |
| Consensus MFE | -34.86 |
| Energy contribution | -35.42 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10762569 105 - 22224390 CAGCCGCAUGCUGGGCAGCGGGAUCUCCAGAGGCGGCAGCGGCUGCAGCCGCUCCCAGUAGCUGUCCCAUAUUGAAGUCAGCUGCCGCAGCGGCGGCGGCAUAGU ..(((((.(((((((((((((((....(((..((((.((((((....)))))).)).))..)))))))....((....)))))))).)))))...)))))..... ( -57.80) >DroSec_CAF1 12260 105 - 1 CAGCCGCAUGCUGGGCAGCGGGAUCUCCAGAGGCGGCAGCGGCUGCAGCCGCACCCAGUAGCUGUCCCAUAUUAAAGUCAGCCGUCGCAGCGGCGGCGGCAUAGU ..(((((....(((((((((((.((....)).(((((.((....)).))))).)))....)))).))))...........(((((....))))).)))))..... ( -46.80) >DroEre_CAF1 10767 105 - 1 CAGCCGCAUGCUGUGCAGCCGGAUCUCCAGAGGCGGCAGCGGCUGCAGCCGCUCCCAGUAGCUGCCCCAUAUUGAAGUCGGCUGCCGCAGCGGCGGCGGCGUAGU ..(((((.((((((((((((((....)).(.((((((((((((....)))))).......)))))).)...........))))).)))))))...)))))..... ( -56.61) >DroWil_CAF1 7617 102 - 1 CAGCUG---GCAUCGACGAUGCGUCGCCAGUGGCAGCAGCAGCUGCCGCAGCUCCCAGCAAUUGUCCCAUAUUGAAGUCAGCUGCCGCAGCAGCGGCUGCAUAAU ..((((---((..((......))..)))))).(((((.((.(((((.((((((..(..((((........))))..)..)))))).))))).)).)))))..... ( -48.50) >DroYak_CAF1 12918 105 - 1 CAGCCGCAUGCUGUGCAGCCGGAUCUCCAGAAGCGGCAGCGGCUGCAGCCGCUCCCAGUAGCUGCCCCAUAUUGAAGUCAGCUGCCGCAGCGGCGGCGGCAUAGU ..(((((.((((((......((....))....((((((((((..(((((..(.....)..))))).))....((....)))))))))).)))))))))))..... ( -51.70) >DroAna_CAF1 20583 105 - 1 CGGCGGCAUGUUGGGCCGCCGGAUCCCCGGAGGCGGCUGCCGCAGCUGCUGCCCCCAAAAGCUGGCUCAUAUUGAAGUCCGCUGACGCGGCGGCGGCAGCAUAGU ..((((((.(((..(((.((((....)))).)))))))))))).((((((((((((...(((.(((((.....).)))).)))...).)).)))))))))..... ( -55.80) >consensus CAGCCGCAUGCUGGGCAGCCGGAUCUCCAGAGGCGGCAGCGGCUGCAGCCGCUCCCAGUAGCUGUCCCAUAUUGAAGUCAGCUGCCGCAGCGGCGGCGGCAUAGU (((((((.((((..((..(.((....)).)..)))))))))))))..(((((.....(((((((.(..........).)))))))(((....))))))))..... (-34.86 = -35.42 + 0.56)

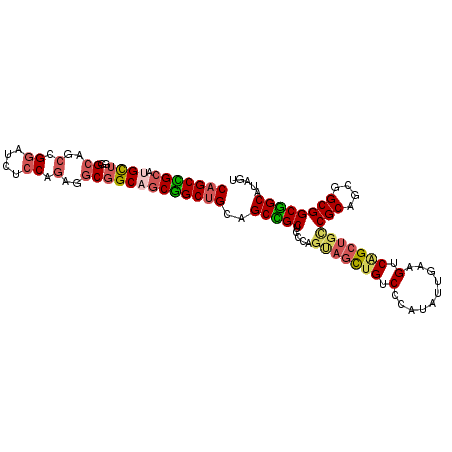

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:32 2006