| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,762,047 – 10,762,164 |
| Length | 117 |
| Max. P | 0.638346 |

| Location | 10,762,047 – 10,762,164 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
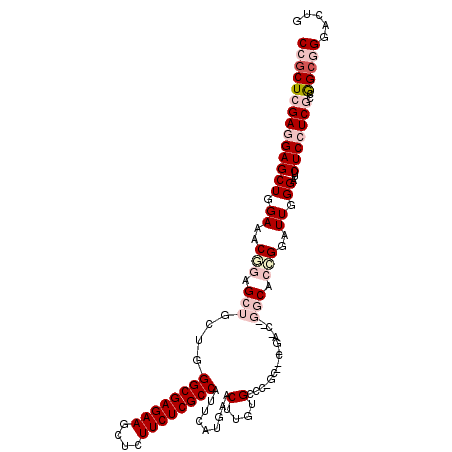
| Mean single sequence MFE | -49.93 |
| Consensus MFE | -28.09 |
| Energy contribution | -30.28 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
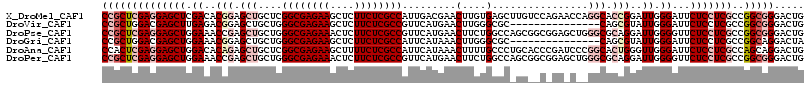
>X_DroMel_CAF1 10762047 117 - 22224390 CCGCUCGAGGAGCUCGACACGGAGCUGCUCGGCGAGAAGCUCUUCUCGCCAUUGACGAACUUGUGAGCUUGUCCAGAACCAGGCACCGGAUUGGGAUUCUCCUCGCCGGCGGGACUG (((((((((((((((((..(((.(((..((((((((((....))))))))...(((((.((....)).)))))..))....))).)))..)))))...)))))))..)))))..... ( -52.80) >DroVir_CAF1 11495 102 - 1 CCGCUGGACGAGCUUGAGACGGAGCUGCUGGGCGAGAAGCUCUUCUCGCCGUUCAUGAACUUGGGCGC---------------CAGCGUAUUGGGAUUCUCCUCGCCGGCGGGACUG (((((((.((((...(((((.((..(((((((((((((....)))))))((((((......)))))))---------------)))))..)).)...)))))))))))))))..... ( -48.40) >DroPse_CAF1 27851 117 - 1 CCGCUCGAGGAGCUGGAAACCGAGCUGCUGGGCGAGAAACUCUUCUCGCCGUUCAUGAACUUCUGGCCAGCGGCGGAGCUGGGCGCAGGAUUGGGGUUCUCCUCGCCGGCGGGACUG ((((((((((((...((..((((.((((..((((((((....))))))))..............(.(((((......))))).)))))..))))..)))))))))..)))))..... ( -56.50) >DroGri_CAF1 11367 102 - 1 CCGCUGGACGAGCUGGAAACGGAGCUGCUGGGCGAGAAGCUCUUCUCGCCAUUCAUAAACUUGGGCGC---------------CAGCGUAUUGGGAUUCUCCUCGCCGGCAGGACUA (((((((.((((..(((..(.((..(((((((((((((....)))))))).((((......))))...---------------)))))..)).)..)))..))))))))).)).... ( -42.50) >DroAna_CAF1 20055 117 - 1 CCACUCGAGGAGCUGGACACAGAGCUGCUCGGCGAGAAGCUUUUCUCGCCAUUCAUAAACUUUUGCCCUGCACCCGAUCCCGGCACUGGGUUGGGAUUCUCCUCGCCAGCAGGACUG ......(((.((((........)))).)))((((((((....))))))))................(((((..(((....))).....(((.(((.....))).))).))))).... ( -42.90) >DroPer_CAF1 20476 117 - 1 CCGCUCGAGGAGCUGGAAACCGAGCUGCUGGGCGAGAAACUCUUCUCGCCGUUCAUGAACUUCUGGCCAGCGGCGGAGCUGGGCGCAGGAUUGGGGUUCUCCUCGCCGGCGGGACUG ((((((((((((...((..((((.((((..((((((((....))))))))..............(.(((((......))))).)))))..))))..)))))))))..)))))..... ( -56.50) >consensus CCGCUCGAGGAGCUGGAAACGGAGCUGCUGGGCGAGAAGCUCUUCUCGCCAUUCAUGAACUUGUGCCC_GC__C_GA_C__GGCACCGGAUUGGGAUUCUCCUCGCCGGCGGGACUG ((((((((((((((.((..(((.(((....((((((((....)))))))).........(....)................))).)))..)).))...)))))))..)))))..... (-28.09 = -30.28 + 2.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:28 2006