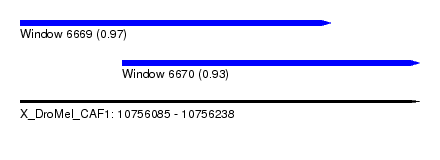
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,756,085 – 10,756,238 |
| Length | 153 |
| Max. P | 0.966068 |

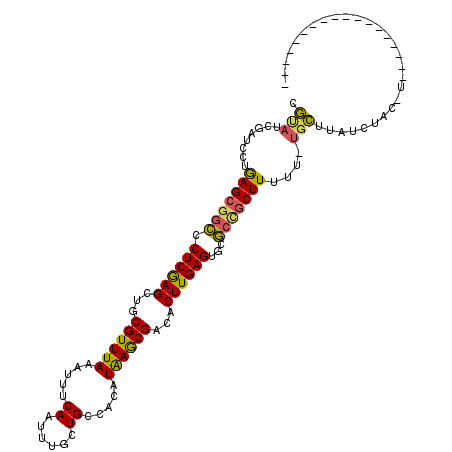
| Location | 10,756,085 – 10,756,204 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -22.55 |
| Energy contribution | -22.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10756085 119 + 22224390 ACCGCAAUAUGCCAUCAUUGCCACUCUCACCUCGA-CACUCGUAUCGAUCCUGAGCGGCCCUCGAGUUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUUGAGUGCGCCGCUU ...(((((........)))))..........((((-........))))....(((((((((((((((..((((((.....((......)).....))))))..))))))).).))))))) ( -31.30) >DroVir_CAF1 3331 91 + 1 ---------------UU-------U------UUUG-CCUUUGCAUCGAUCCUAAGCGGCCCUCAAGCUGCGUUUAAAUUUCAAUUUUCUGCCACAUGAGCGUCACUUGAGUGCGCCGCUU ---------------..-------.------..((-(....)))........((((((((((((((..(((((((.....((......)).....)))))))..)))))).).))))))) ( -27.50) >DroEre_CAF1 4758 114 + 1 ACCA-----UGCCAUCAUUGCCACUCUCACCUCGA-CACUCGUAUCGAUCCUGAGCGGCCCUCGAGUUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUUGAGUGCGCCGCUU ....-----......................((((-........))))....(((((((((((((((..((((((.....((......)).....))))))..))))))).).))))))) ( -28.00) >DroWil_CAF1 2808 102 + 1 --------------UAGAUGGCACCUU---CUUGA-GACACAUAUCGAUCCUGAGCGCUACUCAAGCAGCGUUUAAAUUUCAAUUUGCUGCCACAUAAACGUCACUUGAGUGCACUGCUU --------------.....(((.....---.((((-........)))).....((.((.(((((((..(((((((.....((......)).....)))))))..))))))))).))))). ( -22.60) >DroYak_CAF1 6255 119 + 1 ACCAUAAUAUGCCAUCAUUGCCACUCUCACCUCGA-CACUCGUAUCGAUCCUGAGCGGCCCUCGAGUUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUCGAGUGCGCCGCUU ...............................((((-........))))....(((((((((((((((..((((((.....((......)).....))))))..))))))).).))))))) ( -30.10) >DroAna_CAF1 3548 117 + 1 CGCCUCGAAUGUCCUCAUCAUCA---UCACCUCACCCACUCGUAUCGAUCCUGAGCAGCCCUCGAGCUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUUGAGUGCGCCGCUU ((((((((.((((....(((..(---((..................)))..)))(((((......)))))................(((........))))))).))))).)))...... ( -25.97) >consensus ACC______UGCCAUCAUUGCCACUCUCACCUCGA_CACUCGUAUCGAUCCUGAGCGGCCCUCGAGCUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUUGAGUGCGCCGCUU ....................................................((((((((((((((.((((((((.....((......)).....)))))).)))))))).).))))))) (-22.55 = -22.05 + -0.50)


| Location | 10,756,124 – 10,756,238 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -19.93 |
| Energy contribution | -19.13 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10756124 114 + 22224390 CGUAUCGAUCCUGAGCGGCCCUCGAGUUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUUGAGUGCGCCGCUUUUU-UGCUUAUCUACUUCGCCCCGCCUCUGAUGUC ..(((((.....(((((((((((((((..((((((.....((......)).....))))))..))))))).).)))))))...-.((...........))........))))).. ( -27.20) >DroGri_CAF1 3724 92 + 1 UGCAUCGAACCUAAGCGGCACUCAAGCAGCGUUUAAAUUUCAAUUUUCUGACACAUGAGCGUCACUUGAGUGCGCCGCUU-UU-UGCUUAUCUA--------------------- .(((........((((((((((((((..(((((((....(((......)))....)))))))..)))))))..)))))))-..-))).......--------------------- ( -30.00) >DroEre_CAF1 4792 104 + 1 CGUAUCGAUCCUGAGCGGCCCUCGAGUUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUUGAGUGCGCCGCUU-UU-UGCUUAUCUACUUCGGCCCUCC--------- .(..((((....(((((((((((((((..((((((.....((......)).....))))))..))))))).).)))))))-..-............))))..)...--------- ( -27.47) >DroWil_CAF1 2830 109 + 1 CAUAUCGAUCCUGAGCGCUACUCAAGCAGCGUUUAAAUUUCAAUUUGCUGCCACAUAAACGUCACUUGAGUGCACUGCUUUUU-UCUUUUUUUGUGUUUGCC----AGUGUUA-C ......((..(((.((...(((((((..(((((((.....((......)).....)))))))..)))))))((((........-.........))))..)))----))..)).-. ( -23.03) >DroMoj_CAF1 2468 97 + 1 UGCAUCGAUUCUAAGCGGCCCUCGAGCUGCGUUUAAAUUUCAAUUUUCUGCCACAUGAGCGUCACUUGAGUGCGCCUCUUUUU-UGCUUAUCUGCCAU----------------- .(((........(((.((((((((((..(((((((.....((......)).....)))))))..)))))).).))).)))...-)))...........----------------- ( -22.30) >DroAna_CAF1 3585 106 + 1 CGUAUCGAUCCUGAGCAGCCCUCGAGCUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUUGAGUGCGCCGCUUUUUGCGCUUAUAUAU---------CCUCCGACAUC .((...(((..(((((.((.((((((.((((((((.....((......)).....)))))).)))))))).))...((.....)))))))...))---------).....))... ( -24.00) >consensus CGUAUCGAUCCUGAGCGGCCCUCGAGCUGCGUUUAAAUUUCAAUUUGCUGCCACAUAAGCGACACUUGAGUGCGCCGCUUUUU_UGCUUAUCUAC_U__________________ .(((........(((((((.((((((...((((((.....((......)).....))))))...))))))...)))))))....)))............................ (-19.93 = -19.13 + -0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:19 2006