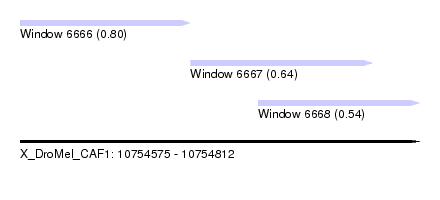
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,754,575 – 10,754,812 |
| Length | 237 |
| Max. P | 0.801779 |

| Location | 10,754,575 – 10,754,676 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -19.66 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10754575 101 + 22224390 CAUUUCCUUU-----CCGGACAACCAGUCCGA------UUCCGAGUAUAAGCCGAUUAGCUGUCUCCCACCUCGUCGAGUUCGAGUUGCUUAGCUGGGCUUGGACUUGGAAU ..........-----.(((((.....)))))(------((((((((.((((((...((((((.....((.((((.......)))).))..)))))))))))).))))))))) ( -36.90) >DroSec_CAF1 4110 96 + 1 CAUUUCCCUUCCAGUCCGGACAAGCAGUCCGA------UUCCAAGUAUAAGCCGAUUAGCUGGCUCCCACCUCGUCGAGUUCGAGUUGCUUAGCUGGGCUUG---------- ............(((((((..((((((..(((------............((((((.((.(((...))).)).)))).)))))..))))))..)))))))..---------- ( -27.70) >DroSim_CAF1 1577 96 + 1 CAUUUCCUUUUCAGUCCGGACAAGCAGUCCGA------UUCCCAGUAUAAGCCGAUUAGCUGUCUCCCACCUCGUCGAGUUCGAGUUGCUUAGCUGGGCUUG---------- ................(((((.....))))).------..((((((.(((((.(((.....)))......((((.......))))..)))))))))))....---------- ( -26.00) >DroEre_CAF1 3391 101 + 1 CACCUCCUCU-----CCGGACAACCAGUCCGA------UUCCGAGUAUAAGCUGAUUAGCUGUCUCCCUCCUCGCCGAGUUCCAGUUGCUCAGCGGGGCUGGGACUGGGAAU ..........-----.(((((.....)))))(------((((.(((...((((....))))....(((.((((((.((((.......)))).))))))..)))))).))))) ( -39.70) >DroYak_CAF1 4766 102 + 1 CACUUCCUUC-----CCGGACAACCAGUCCGAUUCCGAUUCCGAGUAUAAGCUGAUUAGCUGUCUCCUUCCUCGCCGAGUUCCAGUUGCUUGGCGGUGCUUGGACUU----- ..........-----.(((((.....)))))........(((((((((.((((....))))...........((((((((.......)))))))))))))))))...----- ( -34.60) >consensus CAUUUCCUUU_____CCGGACAACCAGUCCGA______UUCCGAGUAUAAGCCGAUUAGCUGUCUCCCACCUCGUCGAGUUCGAGUUGCUUAGCUGGGCUUGGACU______ ................(((((.....)))))........(((((((...(((..(.(((((..(((..........)))....))))).)..)))..)))))))........ (-19.66 = -21.18 + 1.52)



| Location | 10,754,676 – 10,754,784 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -22.84 |
| Energy contribution | -24.77 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10754676 108 + 22224390 GGACGUGGGUUUGGGUUUGAGUUUCGAUCCGGUUGCUGUAGCUAGUGGUGGU---------GGUGGUCUGGGCCAUCGUUUUGCCUUGGCUAAUGAGGGCAACCUGAAACGGACCUC ......(((((((((.(((.....))).))(((((((.(((((((.((..((---------((((((....)))))))..)..))))))))).....))))))).....))))))). ( -42.70) >DroSec_CAF1 4206 109 + 1 -------GGUUUGGGUUUGGGUUUCGAUCUGG-UGCCGUAGCUAGUGGUGGUGGUGUUGGUGGUGGUCUGGGCCACCGUUUUGCCUUGCCUAAUGAGGGCAACCUGAAACGGACCUC -------((((((..((..((((.....(..(-..(((..((.....))..)))..)..).((((((....)))))).....(((((........)))))))))..)).)))))).. ( -41.80) >DroSim_CAF1 1673 100 + 1 -------GGUUUGGGCUUGGGUUUCGAUCUGG-UGCCGUAGCUAGUGGUGGU---------GGUGGUCUGGGCCACCGUUUUGCCUUGGCUAAUGAGGGCAACCUGAAACGGACCUC -------((((((((((..(((....)))..)-.))).(((((((.((..((---------((((((....)))))))..)..))))))))).(.(((....))).)..)))))).. ( -44.00) >DroEre_CAF1 3492 95 + 1 GGGUUG------GGGCUUGGGCUUGGGUGUCG-AACUGGUGGUGCUGC---------------CGGUCUGGGCCACCGUUUUGCCUUGGCUAAUGAGGGCAACCUGAAACGGACCUC (((((.------(((((.((((.(((..(((.-(((((((......))---------------)))).).)))..)))....)))).))))..(.(((....))).)..).))))). ( -38.30) >consensus _______GGUUUGGGCUUGGGUUUCGAUCUGG_UGCCGUAGCUAGUGGUGGU_________GGUGGUCUGGGCCACCGUUUUGCCUUGGCUAAUGAGGGCAACCUGAAACGGACCUC .......((((((((((.((((............(((((.....)))))............((((((....)))))).....)))).))))..(.(((....))).)..)))))).. (-22.84 = -24.77 + 1.94)

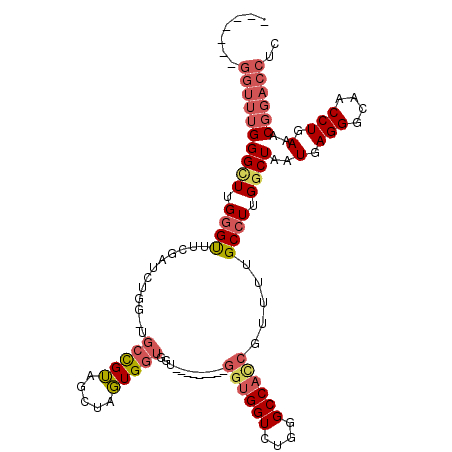
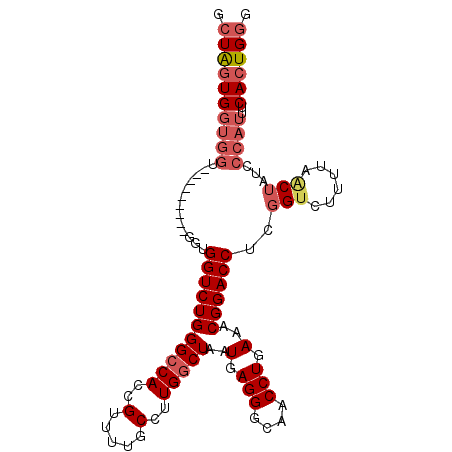
| Location | 10,754,716 – 10,754,812 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -22.99 |
| Energy contribution | -24.93 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10754716 96 + 22224390 GCUAGUGGUGGU---------GGUGGUCUGGGCCAUCGUUUUGCCUUGGCUAAUGAGGGCAACCUGAAACGGACCUCGGUCUUUUAGCUAUCCCAUUUCACUGGG .((((((((((.---------((((((..(((((..((((((((....)).....(((....)))))))))(....))))))....))))))))))..)))))). ( -35.90) >DroSec_CAF1 4238 105 + 1 GCUAGUGGUGGUGGUGUUGGUGGUGGUCUGGGCCACCGUUUUGCCUUGCCUAAUGAGGGCAACCUGAAACGGACCUCGGUCUUUUAACUAUCCCAUUUCACUGGG .(((((((.(((((.(.((((((((((....))))))((((((..((((((.....))))))..))))))((((....))))....)))).))))))))))))). ( -42.40) >DroSim_CAF1 1705 96 + 1 GCUAGUGGUGGU---------GGUGGUCUGGGCCACCGUUUUGCCUUGGCUAAUGAGGGCAACCUGAAACGGACCUCGGUCUUUUAACUAUCCCAUUUCACUGGG .((((((((((.---------((((((..(((((.(((((((((....)).....(((....)))))))))).....)))))....))))))))))..)))))). ( -37.90) >DroEre_CAF1 3525 88 + 1 GGUGCUGC---------------CGGUCUGGGCCACCGUUUUGCCUUGGCUAAUGAGGGCAACCUGAAACGGACCUCUG-CUUUUAACUAUCCCAUUUCACUGG- ((.....)---------------)(((((((((((..(.....)..)))))..(.(((....))).)..))))))....-............(((......)))- ( -24.20) >consensus GCUAGUGGUGGU_________GGUGGUCUGGGCCACCGUUUUGCCUUGGCUAAUGAGGGCAACCUGAAACGGACCUCGGUCUUUUAACUAUCCCAUUUCACUGGG .((((((((((.............(((((((((((..(.....)..)))))..(.(((....))).)..))))))..(((......)))...))))..)))))). (-22.99 = -24.93 + 1.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:18 2006