| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,740,820 – 10,740,928 |
| Length | 108 |
| Max. P | 0.777771 |

| Location | 10,740,820 – 10,740,928 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -18.07 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10740820 108 + 22224390 UUUAAACUAUUCUGUUUCGUUUUAUUUGCCACCACAUGCAUACCUGUUAGUGUCAGUAUAUUGUCGCAGGCAUUUUUGCGCCUACGCGGCACAUCGAUCGGCACAAAG ...((((......)))).........((((..(((..(((....)))..))).........((((((((((........))))..))))))........))))..... ( -22.60) >DroSec_CAF1 16654 99 + 1 CUUAAACCAUUCCAUUUCGUUUUAUUUGCC---------AUACCAGUUAGUGUCAGUAUAUUGUCGCAGGCAUUCUUGCGCCUACGCGGCACAUUGAUCGGCACAAAG ..........................((((---------.(((......)))(((((....((((((((((........))))..)))))).)))))..))))..... ( -21.00) >DroSim_CAF1 6128 108 + 1 CUUAAACCAUUCCAUUUCGUUUUAUUUGCCACCACAUGCAUACCAGUUAGUGUCAGUAUAUUGUCGCAGGCAUUCUUGCGCCUACGCGGCACAUUGAUGGGCACAAAG ..........................((((..(((..((......))..)))(((((....((((((((((........))))..)))))).)))))..))))..... ( -23.10) >DroEre_CAF1 16833 104 + 1 UUAAAACCAUUUUGUUCCGUUUUAUGUGCCACAU----CAUACCCGUCAGUGUUAGUAUAUUGUCGCAGGCAUUCUUGCGCCUACGCGGCACAUCGAUCGGCACAAAG ..........(((((.(((.((.(((((((....----.......(.((((((....)))))).)((((((........))))..))))))))).)).))).))))). ( -32.60) >DroYak_CAF1 16974 104 + 1 UUUAAACCAUUAUGUUUCGUUCCAUGUGCCACAU----CGUACCCGUUAGUGUCAGUAUAUUGUCGCAGGCAUUCUUCCGCCUACGCGGCACAUUGAUCGGCACAAAG ............(((.(((.((.(((((((.(..----...........(((.(((....))).)))((((........))))..).))))))).)).))).)))... ( -28.50) >DroAna_CAF1 19413 104 + 1 UGUGAACUAUUCCUUUAG----AAUGGGCUUCCUUUAGCUUACCUGUGAGUUGAAGUAUAUUGUCGCAGGCAUUUUUCCGCCUGCGAGGCACAUUGACGGGCACGAAC .(((..((((((.....)----)))))(((((((((((((((....)))))))))).........((((((........)))))))))))...........))).... ( -35.90) >consensus UUUAAACCAUUCCGUUUCGUUUUAUGUGCCACCU____CAUACCAGUUAGUGUCAGUAUAUUGUCGCAGGCAUUCUUGCGCCUACGCGGCACAUUGAUCGGCACAAAG .................................................(((((.((....((((((((((........))))..)))))).....)).))))).... (-18.07 = -18.48 + 0.42)

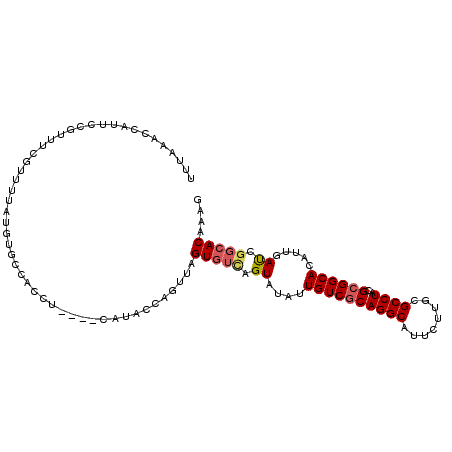

| Location | 10,740,820 – 10,740,928 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.41 |
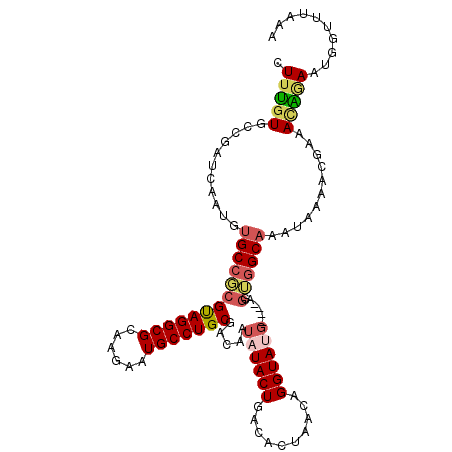
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -20.59 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10740820 108 - 22224390 CUUUGUGCCGAUCGAUGUGCCGCGUAGGCGCAAAAAUGCCUGCGACAAUAUACUGACACUAACAGGUAUGCAUGUGGUGGCAAAUAAAACGAAACAGAAUAGUUUAAA .(((((..((.......(((((((((((((......))))))).(((.((((((..........))))))..))).)))))).......))..))))).......... ( -29.24) >DroSec_CAF1 16654 99 - 1 CUUUGUGCCGAUCAAUGUGCCGCGUAGGCGCAAGAAUGCCUGCGACAAUAUACUGACACUAACUGGUAU---------GGCAAAUAAAACGAAAUGGAAUGGUUUAAG ..(((.((((.((....(((((((((((((......)))))))).)...((((((........))))))---------))))..............)).)))).))). ( -23.97) >DroSim_CAF1 6128 108 - 1 CUUUGUGCCCAUCAAUGUGCCGCGUAGGCGCAAGAAUGCCUGCGACAAUAUACUGACACUAACUGGUAUGCAUGUGGUGGCAAAUAAAACGAAAUGGAAUGGUUUAAG .(((((..((((((.(((((.(((((((((......)))))))).)...((((((........)))))))))))))))))...))))).................... ( -27.60) >DroEre_CAF1 16833 104 - 1 CUUUGUGCCGAUCGAUGUGCCGCGUAGGCGCAAGAAUGCCUGCGACAAUAUACUAACACUGACGGGUAUG----AUGUGGCACAUAAAACGGAACAAAAUGGUUUUAA .(((((.(((....(((((((.((((((((......))))))))(((.((((((..........))))))----.))))))))))....))).))))).......... ( -37.60) >DroYak_CAF1 16974 104 - 1 CUUUGUGCCGAUCAAUGUGCCGCGUAGGCGGAAGAAUGCCUGCGACAAUAUACUGACACUAACGGGUACG----AUGUGGCACAUGGAACGAAACAUAAUGGUUUAAA ((.(((((((..((.(((((((((((((((......)))))))).)............(....)))))))----.))))))))).))....((((......))))... ( -34.00) >DroAna_CAF1 19413 104 - 1 GUUCGUGCCCGUCAAUGUGCCUCGCAGGCGGAAAAAUGCCUGCGACAAUAUACUUCAACUCACAGGUAAGCUAAAGGAAGCCCAUU----CUAAAGGAAUAGUUCACA ....(((..(.((.((((...(((((((((......)))))))))..)))).............((...(((......)))))...----......))...)..))). ( -24.60) >consensus CUUUGUGCCGAUCAAUGUGCCGCGUAGGCGCAAGAAUGCCUGCGACAAUAUACUGACACUAACAGGUAUG____AGGUGGCAAAUAAAACGAAACAGAAUGGUUUAAA .(((((...........(((((((((((((......))))))).....((((((..........))))))......))))))...........))))).......... (-20.59 = -21.07 + 0.47)

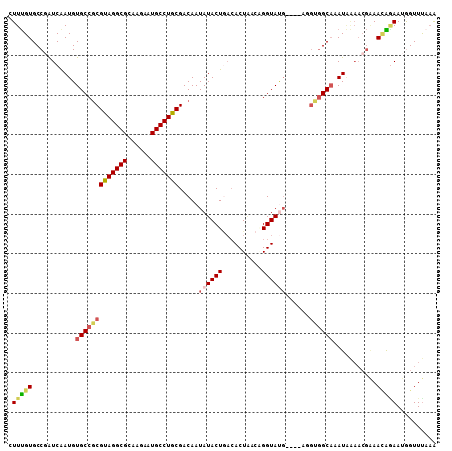
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:11 2006