
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,720,869 – 10,720,979 |
| Length | 110 |
| Max. P | 0.994321 |

| Location | 10,720,869 – 10,720,979 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -20.29 |
| Energy contribution | -21.33 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
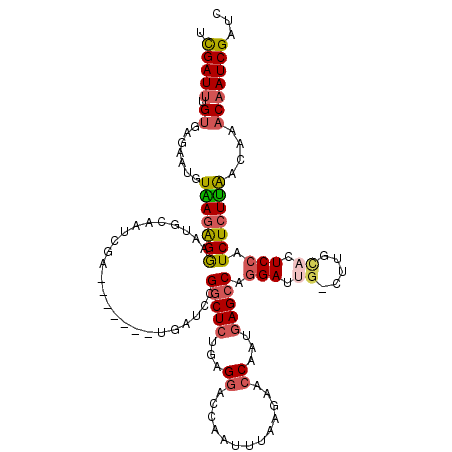
>X_DroMel_CAF1 10720869 110 + 22224390 GAUCGAUUGAUUGUUAAGAGAUGGAGUGCAAG-CAGUCCUGGCUCAUUGGUUCUUAAUUUGGUUCUAAGAGCCGGAUUC-------AAGGUUGCAUUCCUCUUACAUUCUCAC--AUCAA ....((((((.(((...((((.(((((((((.-((((((.(((((.((((..((......))..)))))))))))))).-------..).))))))))))))))))...))).--))).. ( -39.20) >DroSec_CAF1 9668 112 + 1 GAUCGAUUGUUUGUUAAGAGAUGGAGUGCAAG-CAAUCCUGGCUCAUUGGUUCUUAAAUUGGUCCUUAGAGCCUGAUCA-------UCGAUUGCAUUCCUCUUACAUUCUCACAAAUCGA ..((((((((..(....((((.(((((((((.-..(((..(((((...((..(.......)..))...))))).)))..-------....))))))))))))).....)..)).)))))) ( -33.50) >DroSim_CAF1 9699 112 + 1 GAUCGAUUGUUUGUUAAGAGAUGGAGUGCAAG-CAAUCCUGGCUGAUUGGUUCUUAAAUUGGUCCUAAGAGCCGGAUCA-------UCGAUUGCAUUCCUCUUACAUUCUUACAAAUCGA ..((((((((..(....((((.(((((((((.-..((((.((((..((((..((......))..)))).))))))))..-------....))))))))))))).....)..)).)))))) ( -36.50) >DroEre_CAF1 4954 119 + 1 AUACGAUUGUAUGUCAAGAGAUGGAGAUCAAUGCA-UCCUGGCUCAUUGGUUCUUAAAUUGGUCCUCCGAGCCGGAUCAUUACUUAUAGGUUGCAUUUCUCUCACAUUCCCACAAAUCGA ...(((((...(((...(..(((((((..((((((-.((((((((...((..(.......)..))...)))))(........)....))).))))))..)))).)))..).)))))))). ( -32.80) >DroYak_CAF1 9446 119 + 1 AAACGAUUGUAUGCUGAGAGAUGGAGAACAAUGCAAUCGUGGCUCAUUGGUUUCUAAAUUGGUCCUCAGAGACGGAUCAUUAUUUAUCGAUUGCAUUUCCAUCAC-UUCCCACAAAUCGA ...(((((...((..(((.((((((....((((((((((.((((....))))..(((((((((((........))))))..))))).)))))))))))))))).)-))..))..))))). ( -34.90) >consensus GAUCGAUUGUUUGUUAAGAGAUGGAGUGCAAG_CAAUCCUGGCUCAUUGGUUCUUAAAUUGGUCCUAAGAGCCGGAUCA_______UCGAUUGCAUUCCUCUUACAUUCUCACAAAUCGA ...(((((.........((((.(((((((((.......(((((((...((..((......)).))...))))))).((..........)))))))))))))))...........))))). (-20.29 = -21.33 + 1.04)


| Location | 10,720,869 – 10,720,979 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -15.12 |
| Energy contribution | -15.88 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10720869 110 - 22224390 UUGAU--GUGAGAAUGUAAGAGGAAUGCAACCUU-------GAAUCCGGCUCUUAGAACCAAAUUAAGAACCAAUGAGCCAGGACUG-CUUGCACUCCAUCUCUUAACAAUCAAUCGAUC (((((--.(((.....((((((((.(((((.(..-------...((((((((.......................))))).)))..)-.))))).)))...)))))....)))))))).. ( -26.60) >DroSec_CAF1 9668 112 - 1 UCGAUUUGUGAGAAUGUAAGAGGAAUGCAAUCGA-------UGAUCAGGCUCUAAGGACCAAUUUAAGAACCAAUGAGCCAGGAUUG-CUUGCACUCCAUCUCUUAACAAACAAUCGAUC ((((((.((.......((((((((.(((((.(((-------(..((.(((((...((..(.......)..))...))))).))))))-.))))).)))...)))))....)))))))).. ( -28.90) >DroSim_CAF1 9699 112 - 1 UCGAUUUGUAAGAAUGUAAGAGGAAUGCAAUCGA-------UGAUCCGGCUCUUAGGACCAAUUUAAGAACCAAUCAGCCAGGAUUG-CUUGCACUCCAUCUCUUAACAAACAAUCGAUC ((((((((((((((((...((((...((((((..-------((((..((.(((((((.....))))))).)).)))).....)))))-)...).)))))).)))).)))...)))))).. ( -27.10) >DroEre_CAF1 4954 119 - 1 UCGAUUUGUGGGAAUGUGAGAGAAAUGCAACCUAUAAGUAAUGAUCCGGCUCGGAGGACCAAUUUAAGAACCAAUGAGCCAGGA-UGCAUUGAUCUCCAUCUCUUGACAUACAAUCGUAU .(((((((((((((((.((((..((((((.(((...(....).....((((((..((..(.......)..))..))))))))).-))))))..))))))).)))...)))).)))))... ( -34.90) >DroYak_CAF1 9446 119 - 1 UCGAUUUGUGGGAA-GUGAUGGAAAUGCAAUCGAUAAAUAAUGAUCCGUCUCUGAGGACCAAUUUAGAAACCAAUGAGCCACGAUUGCAUUGUUCUCCAUCUCUCAGCAUACAAUCGUUU .(((((((((.((.-(.((((((((((((((((........((.(((........))).)).((((........))))...))))))))))....)))))).)))..)))).)))))... ( -32.00) >consensus UCGAUUUGUGAGAAUGUAAGAGGAAUGCAAUCGA_______UGAUCCGGCUCUGAGGACCAAUUUAAGAACCAAUGAGCCAGGAUUG_CUUGCACUCCAUCUCUUAACAAACAAUCGAUC .(((((.((.......(((((((........................(((((...((.............))...))))).(((.((.....)).))).)))))))....)))))))... (-15.12 = -15.88 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:03 2006