
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,718,727 – 10,718,853 |
| Length | 126 |
| Max. P | 0.605335 |

| Location | 10,718,727 – 10,718,819 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -17.93 |
| Energy contribution | -17.77 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
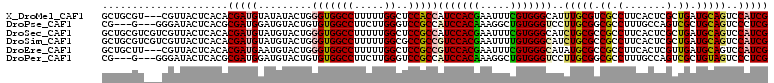
>X_DroMel_CAF1 10718727 92 + 22224390 CCUUCGUU---UGCGACUCU--UGCUGCGU---CGUUACUCACACGAUGUAUAUACUGGGUGGCCUUUUUGGCUCCACCAUCCACGAAUUUCGUGGGCAU ...(((..---..)))....--((((((((---(((.......)))))))).....((((..(((.....)))..).))).(((((.....)))))))). ( -29.00) >DroSec_CAF1 7573 95 + 1 UCUUCGAC---UGUCGCUCU--UGCUGCGUCGUCGUUACUCACACGAUGUAUGUACUGGGUGGCCUUUUUGGCUCCGCCAUCCACGAAUUUCGUGGGCAU ........---.(((((((.--(((((((((((.((.....)))))))))).)))..))))))).....((((...))))((((((.....))))))... ( -32.00) >DroSim_CAF1 7593 95 + 1 CCUUCGAC---UGUCGCUCU--UGCUGCGUCGUCGUUACUCACACGAUGUAUGUACUGGGUGGCCUUUUUGGCGCCGCCGUCCACGAAUUUUGUGGGCAU ........---.(((((((.--(((((((((((.((.....)))))))))).)))..)))))))......(((...)))((((((((...)))))))).. ( -33.10) >DroEre_CAF1 2840 92 + 1 CCAUCGCU---UAGAGCUCU--UGCUGCUU---CGUUACUCACACGAUGAAUGUACUGGGUGGCCUUUUUGGCUCCGCCGUCCACGAAUUUCGUGGGCAU ....((((---((((((...--.)))((((---((((........)))))).)).)))))))(((.....)))......(((((((.....))))))).. ( -30.60) >DroYak_CAF1 7338 97 + 1 CCUUCGGUUAUUAGGGCUCUCAUGCUGCGU---CGUUACUCACACGAUGAAUGUACUGGGUGGCCUUUUUGGCUCCGCCGUCCACGAAUUUCGUGGGCAU .....(((((..(((((((((((((..(((---(((.......))))))...))).)))).))))))..))))).....(((((((.....))))))).. ( -36.90) >DroPer_CAF1 4545 89 + 1 CGUGUGUG---UGUGGCACA--UCG---G---GGGAUACUCACGCGAUGGAUGUACUGUGUGGCCUUCUUGGGUCCGCCAUCCACAAAGGCUGUGGGUCC ((((((.(---(((..(...--...---.---)..)))).))))))..((((.(((.....((((((..(((((.....)))))..))))))))).)))) ( -29.80) >consensus CCUUCGAU___UGUGGCUCU__UGCUGCGU___CGUUACUCACACGAUGUAUGUACUGGGUGGCCUUUUUGGCUCCGCCAUCCACGAAUUUCGUGGGCAU ..........................................................(((((((.....))..)))))(((((((.....))))))).. (-17.93 = -17.77 + -0.17)



| Location | 10,718,745 – 10,718,853 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -42.28 |
| Consensus MFE | -22.94 |
| Energy contribution | -22.75 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10718745 108 + 22224390 GCUGCGU---CGUUACUCACACGAUGUAUAUACUGGGUGGCCUUUUUGGCUCCACCAUCCACGAAUUUCGUGGGCAUUUGCGUCGCCUUCACUCGCUGAUGCAGUCCAUCG (((((((---(...........((((((.....((((..(((.....)))..).)))((((((.....))))))....))))))((........)).))))))))...... ( -36.70) >DroPse_CAF1 4573 105 + 1 CG---G---GGGAUACUCACGCGAUGGAUGUACUGUGUGGCCUUCUUGGGUCCGCCAUCCACAAAGGCUGUGGGUCCUUGCGGCGCCUUUGCCAGUCGCUGCAGUCCCUCG ((---(---(((((((((((((..((((((....(((.(((((....)))))))))))))).....)).))))))...(((((((.((.....)).))))))))))))))) ( -46.70) >DroSec_CAF1 7591 111 + 1 GCUGCGUCGUCGUUACUCACACGAUGUAUGUACUGGGUGGCCUUUUUGGCUCCGCCAUCCACGAAUUUCGUGGGCAUCUGCGCCGCCUUCACUCGCUGAUGCAGUCCAUCG (((((((((((((.......)))))((..((...(((((((.....((((...)))).(((((.....)))))((....)))))))))..))..)).))))))))...... ( -41.50) >DroSim_CAF1 7611 111 + 1 GCUGCGUCGUCGUUACUCACACGAUGUAUGUACUGGGUGGCCUUUUUGGCGCCGCCGUCCACGAAUUUUGUGGGCAUCUGCGCCGCCUUCACUCGCUGAUGCAGUCCAUCG (((((((((((((.......)))))((..((...(((((((......(((...)))((((((((...))))))))......)))))))..))..)).))))))))...... ( -42.10) >DroEre_CAF1 2858 108 + 1 GCUGCUU---CGUUACUCACACGAUGAAUGUACUGGGUGGCCUUUUUGGCUCCGCCGUCCACGAAUUUCGUGGGCAUAUGCGCCGCCUUCACUCGUUGAUGCAGUCCAUCG (((((..---.((.....)).((((((.((....(((((((......(((...)))(((((((.....)))))))......))))))).)).))))))..)))))...... ( -40.00) >DroPer_CAF1 4563 105 + 1 CG---G---GGGAUACUCACGCGAUGGAUGUACUGUGUGGCCUUCUUGGGUCCGCCAUCCACAAAGGCUGUGGGUCCUUGCGGCGCCUUUGCCAGUCGCUGUAGUCCCUCG ((---(---(((((...((.((((((((((....(((.(((((....))))))))))))).....(((...((((((....)).))))..))).)))))))..)))))))) ( -46.70) >consensus GCUGCGU___CGUUACUCACACGAUGUAUGUACUGGGUGGCCUUUUUGGCUCCGCCAUCCACGAAUUUCGUGGGCAUUUGCGCCGCCUUCACUCGCUGAUGCAGUCCAUCG .....................(((((.........(((((((.....))..)))))(((((((.....)))))))..(((((.((.(.......).)).)))))..))))) (-22.94 = -22.75 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:01 2006