| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,670,543 – 10,670,650 |
| Length | 107 |
| Max. P | 0.999735 |

| Location | 10,670,543 – 10,670,650 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -29.11 |
| Energy contribution | -29.33 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
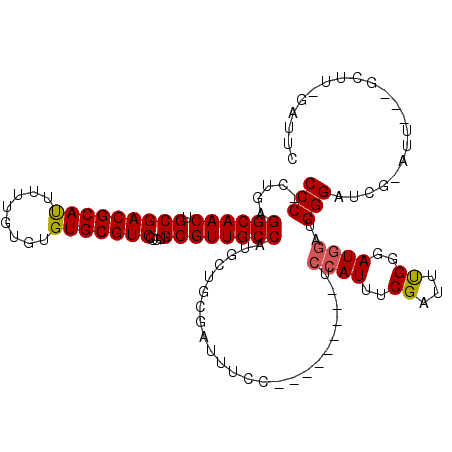
>X_DroMel_CAF1 10670543 107 + 22224390 CCCCCAGCGGCAACUGCGACGCAUUUUUGUGUGUGCGUCCUGUGCGUUGCCAUGCUGCGAUUU-----------UCCAUUUCGAUUUGGGAUGGUUGGGAUCGCAUU--GGCUUGGAUUC ..((.(((((((((.(((((((((........)))))))....))))))))....((((((((-----------.((((..(.....)..))))...))))))))..--.))).)).... ( -43.80) >DroEre_CAF1 835 89 + 1 CCC-CUGAGGCAACUGCGACGCAUUUUUGUGUGUGCGUCCUGUGCGUUGCCAUGCUGCGAUUUCC------------AUUUCGAUUCGGGAUGGAUGGGUUC------------------ .((-(...((((((.(((((((((........)))))))....))))))))...........(((------------((..((...))..))))).)))...------------------ ( -34.80) >DroYak_CAF1 1854 119 + 1 CCC-CUGAGGCAACUGCGACGCACUUUUGUGUGUGCGUCCUGUGCGUUGCCAUGCUGCGAUUUCCAUUUCCAUUUCCAUUUCGAUUUGGGAUGGCUGGGAUCGUAUUCUCGCUUAGAUUC ...-((((((((((.(((((((((........)))))))....))))))))....(((((((.(((...(((((.(((........)))))))).)))))))))).......)))).... ( -41.90) >consensus CCC_CUGAGGCAACUGCGACGCAUUUUUGUGUGUGCGUCCUGUGCGUUGCCAUGCUGCGAUUUCC_________UCCAUUUCGAUUUGGGAUGGAUGGGAUCG_AUU___GCUU_GAUUC (((.....((((((.(((((((((........)))))))....))))))))........................((((..((...))..))))..)))..................... (-29.11 = -29.33 + 0.23)


| Location | 10,670,543 – 10,670,650 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -21.67 |
| Energy contribution | -22.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
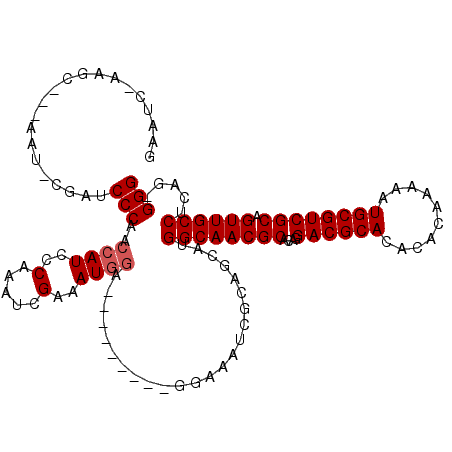
>X_DroMel_CAF1 10670543 107 - 22224390 GAAUCCAAGCC--AAUGCGAUCCCAACCAUCCCAAAUCGAAAUGGA-----------AAAUCGCAGCAUGGCAACGCACAGGACGCACACACAAAAAUGCGUCGCAGUUGCCGCUGGGGG ...(((.(((.--..((((((.....((((..(.....)..)))).-----------..))))))....((((((((....((((((..........)))))))).))))))))).))). ( -33.40) >DroEre_CAF1 835 89 - 1 ------------------GAACCCAUCCAUCCCGAAUCGAAAU------------GGAAAUCGCAGCAUGGCAACGCACAGGACGCACACACAAAAAUGCGUCGCAGUUGCCUCAG-GGG ------------------...(((.(((((..((...))..))------------)))...........((((((((....((((((..........)))))))).))))))...)-)). ( -28.10) >DroYak_CAF1 1854 119 - 1 GAAUCUAAGCGAGAAUACGAUCCCAGCCAUCCCAAAUCGAAAUGGAAAUGGAAAUGGAAAUCGCAGCAUGGCAACGCACAGGACGCACACACAAAAGUGCGUCGCAGUUGCCUCAG-GGG ...(((..((((((......))(((.((((.(((........)))..))))...)))...)))).....((((((((....(((((((........))))))))).))))))..))-).. ( -35.70) >consensus GAAUC_AAGC___AAU_CGAUCCCAACCAUCCCAAAUCGAAAUGGA_________GGAAAUCGCAGCAUGGCAACGCACAGGACGCACACACAAAAAUGCGUCGCAGUUGCCUCAG_GGG .....................(((..((((..(.....)..))))........................((((((((....((((((..........)))))))).)))))).....))) (-21.67 = -22.33 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:48 2006