
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,654,070 – 10,654,160 |
| Length | 90 |
| Max. P | 0.877171 |

| Location | 10,654,070 – 10,654,160 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -10.62 |
| Energy contribution | -10.99 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
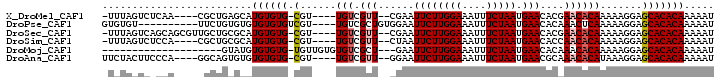
>X_DroMel_CAF1 10654070 90 + 22224390 -UUUAGUCUCAA----CGCUGAGCAUGUGUG-CGU----UGUCGUU--CGAAUUCUUGGAAAUUUCUAAUGAACACGAACACAAAAAGGAGCACACAAAAAU -....(.((((.----...))))).((((((-(.(----(((.(((--((..((((((((....))))).)))..)))))))))......)))))))..... ( -26.70) >DroPse_CAF1 13671 88 + 1 GUGUGU----------UUCUGUGUGUGUGUGUCGU----UGUCGCUGUGGAAUUCUUGGAAAUUUCUAAUGAACACAAACUCAAAAAGGAGCACACAAAAAU ((((((----------((((.((.((.((((((((----((.......((((((......))))))))))).))))).)).))...))))))))))...... ( -26.61) >DroSec_CAF1 11716 94 + 1 -UUUAGUCAGCAGCGUUGCUGCGCAUGUGUG-CGU----UGUCGUU--CGAAUUCUUGGAAAUUUCUAAUGAACACGAACACAAAAAGGAGCACACAAAAAU -........(((((...)))))...((((((-(.(----(((.(((--((..((((((((....))))).)))..)))))))))......)))))))..... ( -30.90) >DroSim_CAF1 9800 90 + 1 -UUUAGUCUCCA----CGCUGCGCAUGUGUG-CGU----UGUCGUU--CUAAUUCUUGGAAAUUUCUAAUGAACACCAACACAAAAAGGAGCACACAAAAAU -....((((((.----....((((....)))-)((----((..(((--(......(((((....))))).))))..)))).......)))).))........ ( -21.60) >DroMoj_CAF1 13744 78 + 1 --------------------GUAUGUGUGUG-UGUUGUGUGUCGCU---GAAUUCUUGGAAAUUUCUAAUGAACACAAACACAAAAAGGAGCACACAAAAAU --------------------...(((((((.-..((((((.....(---(..((((((((....))))).)))..)).))))))......)))))))..... ( -18.80) >DroAna_CAF1 12787 91 + 1 UUCUACUUCCCA----GGCAGUGUGUGUGUG-CGU----UGUCGUU--GGAAUUCUUGGAAAUUUCUAAUGAACGCAAACACAUAAAGGAGCACACAAAAAU ......((((..----.....(((((((.((-(((----..(((((--((((...........)))))))))))))).)))))))..))))........... ( -24.50) >consensus _UUUAGUC_CCA____CGCUGUGCAUGUGUG_CGU____UGUCGUU__CGAAUUCUUGGAAAUUUCUAAUGAACACAAACACAAAAAGGAGCACACAAAAAU .........................((((((.(......(((.(((......((((((((....))))).)))....)))))).......)))))))..... (-10.62 = -10.99 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:43 2006