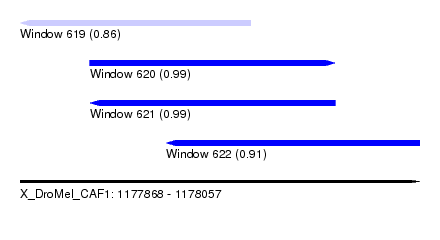
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,177,868 – 1,178,057 |
| Length | 189 |
| Max. P | 0.994146 |

| Location | 1,177,868 – 1,177,977 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -18.57 |
| Energy contribution | -19.21 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177868 109 - 22224390 GAUACAAAUCUCUCUCGAGUGGACACUUAACAUGCGAGGAAGAGAUGGAGAGAGCAAGAGAGAGGGAGGGUGAGAA-----------GGGAGCGACUUGAAUUCUUCCUUGGCGCCCAUG ........((((((((..((.....(((..(((.(........))))..))).))..))))))))..(((((..((-----------(((((...........)))))))..)))))... ( -33.30) >DroSec_CAF1 13752 109 - 1 GAAACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGAAGAGAUGGAGUGAGCGAGAGAGAGGGAGGGUGAGAA-----------GGGAGCGACUUAAAUUCAUUUUUGGCGCCCAUG ........((((((((..((...(((((......(......).....))))).))..))))))))..(((((...(-----------((((..((.......)).)))))..)))))... ( -30.30) >DroSim_CAF1 28824 109 - 1 GAAACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGAAGAGAUGGAGUGAGCGAGAGAGAGGGAGGGUGAGAA-----------GGGAGCGACUUAAAUUCCUUUUUGGCGCCCAUG ....((..(((..((((.(((.........))).))))..)))..))..(((.(((...(((((((((..((((..-----------(....)..))))..)))))))))..))).))). ( -35.80) >DroEre_CAF1 13113 96 - 1 GAUACGAAUCUCUCUCGAGUGGGCACUCAACAUGCGAGGCAGAG-------------GAGAGAGAGAGGGUGCGAA-----------GGGAGCGACUUGAACUCCUUUUUGGCGCCCAUG ........((((((((......((.(((.......)))))....-------------..))))))))(((((((((-----------(((((.........))))))))..))))))... ( -38.90) >DroYak_CAF1 15950 103 - 1 GAUACAAAUCUCUCUCGAGUGGGCACUCAACAUGCGAGGCAGCG--------------AGACAGCGAGGGAGAGGAGCAGAGGAGGAGAGAGAG---GGAACUCCUUUUUGGCGCCCAUG ........((((((((......((.(((.......))))).((.--------------.....))))))))))((.((.(((((((((......---....))))))))).))..))... ( -34.90) >consensus GAUACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGAAGAGAUGGAG_GAGC_AGAGAGAGGGAGGGUGAGAA___________GGGAGCGACUUGAAUUCCUUUUUGGCGCCCAUG ........((((((((((((....))))......(......).................))))))))(((((...............((......))...............)))))... (-18.57 = -19.21 + 0.64)

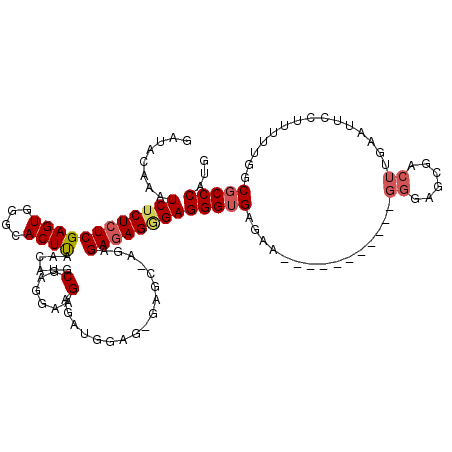
| Location | 1,177,901 – 1,178,017 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -44.74 |
| Consensus MFE | -33.18 |
| Energy contribution | -33.26 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177901 116 + 22224390 ----UUCUCACCCUCCCUCUCUCUUGCUCUCUCCAUCUCUUCCUCGCAUGUUAAGUGUCCACUCGAGAGAGAUUUGUAUCUCAAGGACGCACUUAAAGGAGGCGCGAGUGGGAGGGCAGC ----........(((((((((.(((((.(.((((........(......)....((((((.(((....)))..(((.....))))))))).......))))).))))).))))))).)). ( -41.80) >DroSec_CAF1 13785 116 + 1 ----UUCUCACCCUCCCUCUCUCUCGCUCACUCCAUCUCUUCCUCGCAUGUUAAGUGCCCACUCGAGAGAGAUUUGUUUCUCAAGGACGCACUUAAAGGAGGCGCGAGUGGGAGGGCAAC ----......(((((((.....(((((.(.((((........(......)((((((((...((.((((((......)))))).))...)))))))).))))).))))).))))))).... ( -45.50) >DroSim_CAF1 28857 116 + 1 ----UUCUCACCCUCCCUCUCUCUCGCUCACUCCAUCUCUUCCUCGCAUGUUAAGUGCCCACUCGAGAGAGAUUUGUUUCUCAAGGACGCACUUAAAGGAGGCGCGAGUGGGAGGGCAGC ----........(((((((((.(((((.(.((((........(......)((((((((...((.((((((......)))))).))...)))))))).))))).))))).))))))).)). ( -45.90) >DroEre_CAF1 13146 103 + 1 ----UUCGCACCCUCUCUCUCUC-------------CUCUGCCUCGCAUGUUGAGUGCCCACUCGAGAGAGAUUCGUAUCUCAAGGACGCACUUAAAGGAGGCGCGAGUGGGAGGGCAGC ----...((.(((((((.(((..-------------...((((((.(...((((((((...((.((((..........)))).))...)))))))).))))))).))).)))))))..)) ( -40.80) >DroYak_CAF1 15987 106 + 1 CUGCUCCUCUCCCUCGCUGUCU--------------CGCUGCCUCGCAUGUUGAGUGCCCACUCGAGAGAGAUUUGUAUCUCAAGGACGCACUUAAAGGAGGCGCGAGUGGGAGGGCAGC ((((.((((((.(((((.((((--------------(((......))...((((((((((.(((....)))..(((.....)))))..))))))))..)))))))))).)))))))))). ( -49.70) >consensus ____UUCUCACCCUCCCUCUCUCU_GCUC_CUCCAUCUCUUCCUCGCAUGUUAAGUGCCCACUCGAGAGAGAUUUGUAUCUCAAGGACGCACUUAAAGGAGGCGCGAGUGGGAGGGCAGC ..........(((((((.(((....................((((.(...((((((((((.(((....)))..(((.....)))))..)))))))).)))))...))).))))))).... (-33.18 = -33.26 + 0.08)

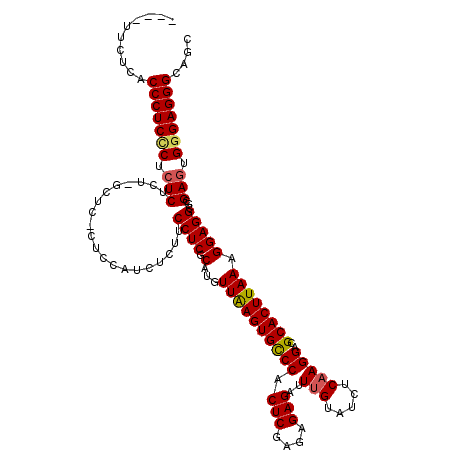
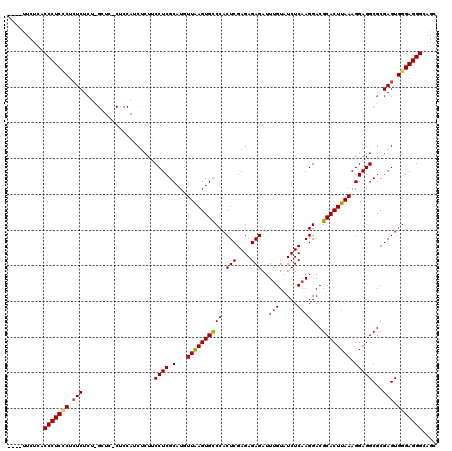
| Location | 1,177,901 – 1,178,017 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -35.66 |
| Energy contribution | -37.06 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177901 116 - 22224390 GCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAUACAAAUCUCUCUCGAGUGGACACUUAACAUGCGAGGAAGAGAUGGAGAGAGCAAGAGAGAGGGAGGGUGAGAA---- .((((((((((.(((((((((......)).)))).((((........((((((((((.(((.........))).))))..))))))........)))).))).)))))))).))..---- ( -43.89) >DroSec_CAF1 13785 116 - 1 GUUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAAACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGAAGAGAUGGAGUGAGCGAGAGAGAGGGAGGGUGAGAA---- .(..(((((((.(((.(((((((.((((((((.(.(((((((..........))))))).).))))))))....(......)....))))...)))...))).)))))))..)...---- ( -49.50) >DroSim_CAF1 28857 116 - 1 GCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAAACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGAAGAGAUGGAGUGAGCGAGAGAGAGGGAGGGUGAGAA---- .((((((((((.(((.(((((((.((((((((.(.(((((((..........))))))).).))))))))....(......)....))))...)))...))).)))))))).))..---- ( -48.20) >DroEre_CAF1 13146 103 - 1 GCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAUACGAAUCUCUCUCGAGUGGGCACUCAACAUGCGAGGCAGAG-------------GAGAGAGAGAGGGUGCGAA---- ((.((((((.(.(((.(((((((.((.(((((.(.(((((((..........))))))).).))))).))...).))))).)..-------------..))).).))))))))...---- ( -40.00) >DroYak_CAF1 15987 106 - 1 GCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAUACAAAUCUCUCUCGAGUGGGCACUCAACAUGCGAGGCAGCG--------------AGACAGCGAGGGAGAGGAGCAG .((((((((((.(((((((((((.((.(((((.(.(((((((..........))))))).).))))).))...).))))).(..--------------...).))))))).)))).)))) ( -46.80) >consensus GCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAUACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGAAGAGAUGGAG_GAGC_AGAGAGAGGGAGGGUGAGAA____ ...((((((((.(((...(((((.((((((((.(.(((((((..........))))))).).))))))))...).))))....................))).))))))))......... (-35.66 = -37.06 + 1.40)

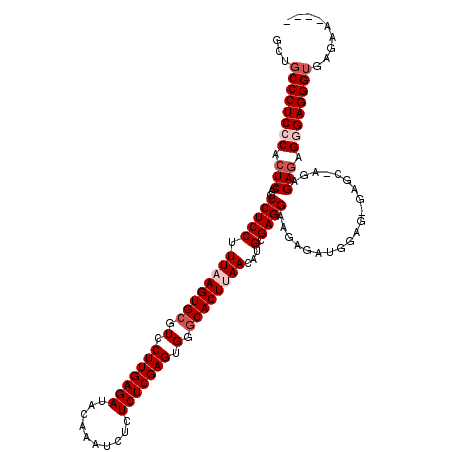
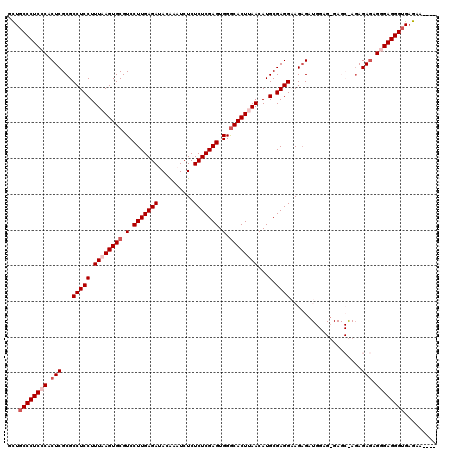
| Location | 1,177,937 – 1,178,057 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -35.16 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177937 120 - 22224390 UCAUAAGCGUGACUCACGGCCAUGACCCCGUCCAAAUGUGGCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAUACAAAUCUCUCUCGAGUGGACACUUAACAUGCGAGGA ......(((((.....((((((((((...))).....))))))).....((((((((((((......)).))))....(((((....)))))....))))))........)))))..... ( -35.80) >DroSec_CAF1 13821 120 - 1 UCAUAUGCGUGACUCACGGCCAUGACUCCGUCCAAAUGUGGUUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAAACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGA ......((((((...((((........))))......((((........))))))))))((((.((((((((.(.(((((((..........))))))).).))))))))...).))).. ( -36.70) >DroSim_CAF1 28893 120 - 1 UCAUAAGCGUGACUCACGGCCAUGACCCCGUCCAAAUGUGGCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAAACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGA ......(((((.....((((((((((...))).....)))))))....(((((((((((((......)).))))....((((......))))....))))))).......)))))..... ( -37.10) >DroEre_CAF1 13169 120 - 1 UCAUAAGCGUGACUCACGGCCAUGACCCCGUGCAAAUGUGGCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAUACGAAUCUCUCUCGAGUGGGCACUCAACAUGCGAGGC ......(((((.....(((((((..............)))))))....(((((((((((((......)).))))....(((((....)))))....))))))).......)))))..... ( -37.64) >DroYak_CAF1 16013 120 - 1 UCAUAAGCGUGACUCACGGCCAUGACCCCGUGCAAAUGUGGCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAUACAAAUCUCUCUCGAGUGGGCACUCAACAUGCGAGGC ......(((((.....(((((((..............)))))))....(((((((((((((......)).))))....(((((....)))))....))))))).......)))))..... ( -37.64) >consensus UCAUAAGCGUGACUCACGGCCAUGACCCCGUCCAAAUGUGGCUGCCCUCCCACUCGCGCCUCCUUUAAGUGCGUCCUUGAGAUACAAAUCUCUCUCGAGUGGGCACUUAACAUGCGAGGA ......(((((.....(((((((..............)))))))....(((((((((((((......)).))))....(((((....)))))....))))))).......)))))..... (-35.16 = -35.60 + 0.44)

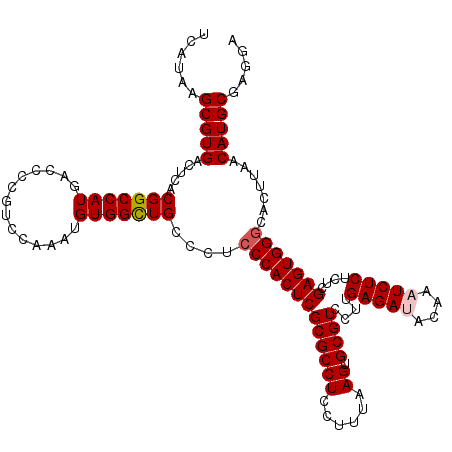
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:47 2006