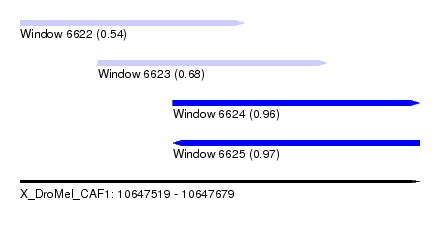
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,647,519 – 10,647,679 |
| Length | 160 |
| Max. P | 0.971862 |

| Location | 10,647,519 – 10,647,609 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 75.95 |
| Mean single sequence MFE | -15.17 |
| Consensus MFE | -2.10 |
| Energy contribution | -2.94 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.14 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10647519 90 + 22224390 UCACUACGGCGCCCUGCCUCUGUCCCACUCACACUCAUUAGGCACUAGUCAGUGCAACAAUUUUACAUUUU-UG------UUUUUGUUUUA----UGCCGU .....(((((((.(((.((..((.((..............)).)).)).))).))(((((....(((....-))------)..)))))...----.))))) ( -18.64) >DroSec_CAF1 4892 84 + 1 UCACCACGGCGCCCUACCUCUGUCCCACUCACACUCAUUAGCCACAAGUCUGUGCAACAAUUAUACAUUUU-U------------GUUUUG----UGCCGU .....(((((((............................((.(((....)))))(((((..........)-)------------)))..)----)))))) ( -14.60) >DroEre_CAF1 5085 90 + 1 UCACCACGGCGCCCUGUCUCUGUCUCACUCACACUCAUUAACCACUAGUCUGUGCAACAAUUAUACAGUUU-UG------UUUUUGUUUUG----UGCUGU .....(((((((...................(((..((((.....))))..))).(((((..(((......-))------)..)))))..)----)))))) ( -13.60) >DroYak_CAF1 5180 90 + 1 UCACCACGGCGCCCUAUCUCUGUCUCACUCACACUCAUUAACAACUAGUGUGUGCAACAAUUUUACAUUUU-UG------UUUUUGUUUUG----UGCUGU .....(((((((...................(((.(((((.....))))).))).(((((....(((....-))------)..)))))..)----)))))) ( -17.60) >DroAna_CAF1 5458 89 + 1 UU-------CGCCCUCUCUCUGUCU-ACU-AAAGUCAUUAACC---AGACUGUGCAACAAUUUCACCUUUCUUGUGCUUCUUCUUGGUCUGUUACUGUUGU ..-------................-((.-..(((.((..(((---(((..(..(((.((........)).)))..)....)).))))..)).)))...)) ( -11.40) >consensus UCACCACGGCGCCCUAUCUCUGUCUCACUCACACUCAUUAACCACUAGUCUGUGCAACAAUUUUACAUUUU_UG______UUUUUGUUUUG____UGCUGU .....((((((......)..(((..(((.......................)))..))).....................................))))) ( -2.10 = -2.94 + 0.84)

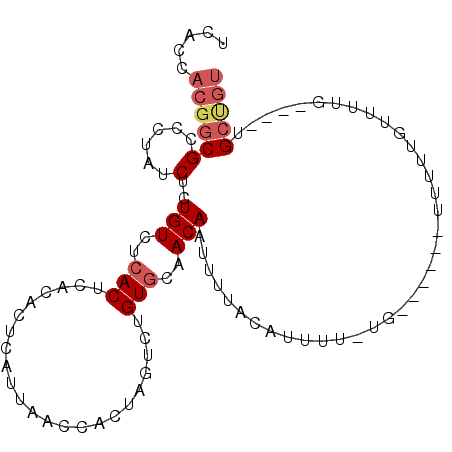

| Location | 10,647,550 – 10,647,642 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.66 |
| Mean single sequence MFE | -21.12 |
| Consensus MFE | -13.59 |
| Energy contribution | -15.15 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10647550 92 + 22224390 CACUCAUUAGGCACUAGUCAGUGCAACAAUUUUACAUUUUUGUUUUUGUUUUAUGCCGUGGUUGUUUUCAUAUAACCAGGUGUGCACACAUG------- .........(((....))).(((((((((....(((....)))..)))))..(((((.(((((((......)))))))))))))))).....------- ( -23.20) >DroSec_CAF1 4923 86 + 1 CACUCAUUAGCCACAAGUCUGUGCAACAAUUAUACAUUUUU------GUUUUGUGCCGUGGUUGUUUUCAUAUAACCAGGUGUGCACACAUG------- ................((.((((((((((....(((....)------)).))))(((.(((((((......)))))))))).)))))).)).------- ( -21.00) >DroEre_CAF1 5116 99 + 1 CACUCAUUAACCACUAGUCUGUGCAACAAUUAUACAGUUUUGUUUUUGUUUUGUGCUGUGGUUGUUUUCAUUUUACCAGGUGUGCACACAUGUACAUAU .......(((((((......(..((((((..(((......)))..))))..))..).)))))))...............(((((((....))))))).. ( -19.90) >DroYak_CAF1 5211 95 + 1 CACUCAUUAACAACUAGUGUGUGCAACAAUUUUACAUUUUUGUUUUUGUUUUGUGCUGUGGUUGUUUUCAUUUUACCAGGUGUGCACACAUGU----AU ................(((((((((((((....(((....)))..)))))..(..((.((((............))))))..)))))))))..----.. ( -20.40) >consensus CACUCAUUAACCACUAGUCUGUGCAACAAUUAUACAUUUUUGUUUUUGUUUUGUGCCGUGGUUGUUUUCAUAUAACCAGGUGUGCACACAUG_______ ....................(((((((((....(((....)))..)))))..(..((.(((((((......)))))))))..)))))............ (-13.59 = -15.15 + 1.56)

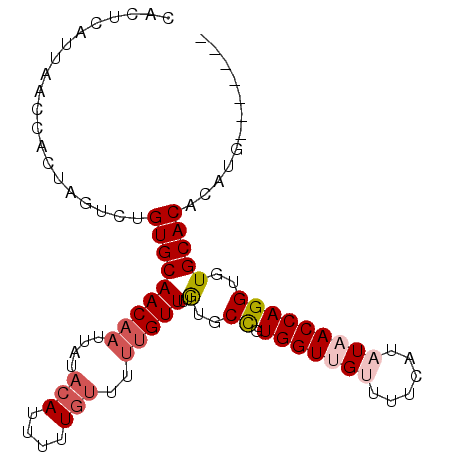
| Location | 10,647,580 – 10,647,679 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10647580 99 + 22224390 UUUACAUUUUUGUUUUUGUUUUAUGCCGUGGUUGUUUUCAUAUAACCAGGUGUGCACACAUG----------UGUAUGUGUGAAUGUUGUUUCUUUUACCUGACAUGUG .....................((((((.(((((((......)))))))))))))..((((((----------(.((.(((.(((......)))...))).))))))))) ( -22.60) >DroSec_CAF1 4953 93 + 1 UAUACAUUUUU------GUUUUGUGCCGUGGUUGUUUUCAUAUAACCAGGUGUGCACACAUG----------UGCAUGUGUGAAUGUUGUUUCUUUUACCUGACAUGUG ...........------....((..((.(((((((......))))))).).)..))((((((----------(.((.(((.(((......)))...))).))))))))) ( -26.10) >DroEre_CAF1 5146 109 + 1 UAUACAGUUUUGUUUUUGUUUUGUGCUGUGGUUGUUUUCAUUUUACCAGGUGUGCACACAUGUACAUAUGGGUGCAUGUGUGUAUGUUGUUCCUUUUACCUGACAUGUG ..((((((................))))))................(((((((((((((((((((......))))))))))))))............)))))....... ( -29.79) >consensus UAUACAUUUUUGUUUUUGUUUUGUGCCGUGGUUGUUUUCAUAUAACCAGGUGUGCACACAUG__________UGCAUGUGUGAAUGUUGUUUCUUUUACCUGACAUGUG ......................(..((.(((((((......)))))))))..).((((((((............)))))))).(((((.............)))))... (-19.31 = -19.32 + 0.01)

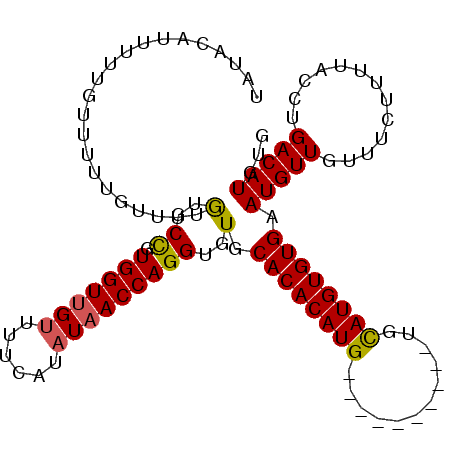

| Location | 10,647,580 – 10,647,679 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -13.47 |
| Energy contribution | -14.13 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10647580 99 - 22224390 CACAUGUCAGGUAAAAGAAACAACAUUCACACAUACA----------CAUGUGUGCACACCUGGUUAUAUGAAAACAACCACGGCAUAAAACAAAAACAAAAAUGUAAA .((((..(((((....(((......)))((((((...----------.))))))....)))))(((.((((....(......).)))).)))..........))))... ( -16.40) >DroSec_CAF1 4953 93 - 1 CACAUGUCAGGUAAAAGAAACAACAUUCACACAUGCA----------CAUGUGUGCACACCUGGUUAUAUGAAAACAACCACGGCACAAAAC------AAAAAUGUAUA ....((((((((....(((......))).....((((----------(....))))).))))((((..........))))..))))......------........... ( -16.10) >DroEre_CAF1 5146 109 - 1 CACAUGUCAGGUAAAAGGAACAACAUACACACAUGCACCCAUAUGUACAUGUGUGCACACCUGGUAAAAUGAAAACAACCACAGCACAAAACAAAAACAAAACUGUAUA ....(..(((((....(......)...((((((((.((......)).))))))))...)))))..)..............((((..................))))... ( -19.47) >consensus CACAUGUCAGGUAAAAGAAACAACAUUCACACAUGCA__________CAUGUGUGCACACCUGGUUAUAUGAAAACAACCACGGCACAAAACAAAAACAAAAAUGUAUA ..((((((((((....(......)...((((((((............))))))))...))))))...))))...................................... (-13.47 = -14.13 + 0.67)

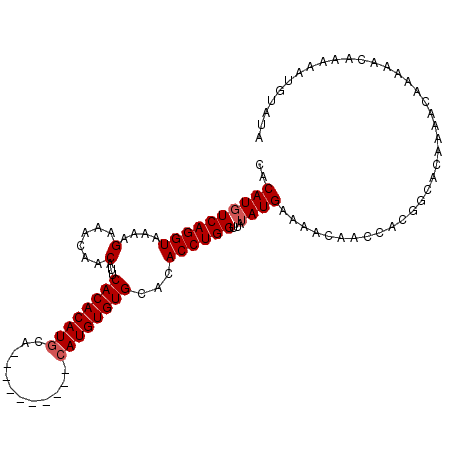
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:40 2006