
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,643,369 – 10,643,529 |
| Length | 160 |
| Max. P | 0.988047 |

| Location | 10,643,369 – 10,643,489 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -51.93 |
| Consensus MFE | -43.86 |
| Energy contribution | -44.37 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
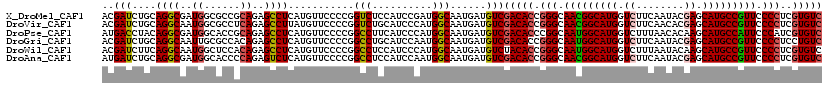
>X_DroMel_CAF1 10643369 120 + 22224390 GGCACCACACUAGCCGGAGUCUUGGACAUGGCAAAGCUUUGACACGAGGGGAACGGCAUGCUCGUAUUGAAGACCAUGCCGUUGCCCGGUGUCGACAUCAUUGCCAUCGGAUGGAGACCG (((.........))).(.(((((.(.((((((((.(..(((((((..(((.(((((((((.((........)).))))))))).))).)))))))...).))))))).)..).)))))). ( -52.20) >DroVir_CAF1 890 120 + 1 GGCACCACGCUGGCCGGUGUCUUGGACAUGGCAAAGCUUUGACACGAGGGGAACGGCAUGCUCGUGUUGAAGACCAUGCCGUUGCCCGGUGUCGACAUCAUUGCCAUGGGAUGCAGACCG .((((((((((....)))))......((((((((.(..(((((((..(((.(((((((((.((........)).))))))))).))).)))))))...).)))))))))).)))...... ( -56.20) >DroPse_CAF1 718 120 + 1 GGAACUACGCUGGCCGGCGUCUUGGACAUGGCAAAGCUCUGACACGAUGGGAAUGGCAUGCUUGUGUUAAAGACCAUGCCAUUGCCGGGUGUCGACAUCAUUGCCAUGGGAUGAAGGCCG ...........((((..(((((....((((((((.(...((((((..(((.((((((((((((......)))..))))))))).))).))))))....).)))))))))))))..)))). ( -52.60) >DroGri_CAF1 12163 120 + 1 GGCACCACGCUGGCCGGUGUUUUGGACAUGGCAAAGCUUUGACAGGAGGGGAACGGCAUGCUCGUAUUGAAGACCAUGCCAUUGCCCGGUGUCGACAUCAUUGCCAUUGGAUGCAGGCCG (((.....)))((((.((((((.....(((((((.(..((((((...(((.((.((((((.((........)).)))))).)).)))..))))))...).))))))).)))))).)))). ( -47.20) >DroWil_CAF1 544 120 + 1 GGCACAACGUUCGCCGGCGUCUUAGACAUGGCAAAGCCCUGACACGAGGGGAACGGCAUGCUUGUAUUAAAGACCAUGCCAUUGCCCGGUGUAGACAUCAUUGCCAUGGGAUGGAGGCCG (((.........)))(((.(((....((((((((.......((((..(((.((.(((((((((......)))..)))))).)).))).))))........))))))))....))).))). ( -49.06) >DroAna_CAF1 691 120 + 1 GGCACCACACUGGCCGGUGUUUUGGACAUGGCAAAACUUUGACACGAGGGGAACGGCAUGCUCGUAUUGAAGACCAUGCCGUUGCCCGGUGUCGACAUCAUUGCCAUUGGAUGGAGGCCG (((.(((((((....)))))......((((((((....(((((((..(((.(((((((((.((........)).))))))))).))).))))))).....)))))).))...))..))). ( -54.30) >consensus GGCACCACGCUGGCCGGUGUCUUGGACAUGGCAAAGCUUUGACACGAGGGGAACGGCAUGCUCGUAUUGAAGACCAUGCCAUUGCCCGGUGUCGACAUCAUUGCCAUGGGAUGGAGGCCG (((.........)))(((.(.(((..((((((((....(((((((..(((.(((((((((.((........)).))))))))).))).))))))).....))))))).)..)))).))). (-43.86 = -44.37 + 0.50)

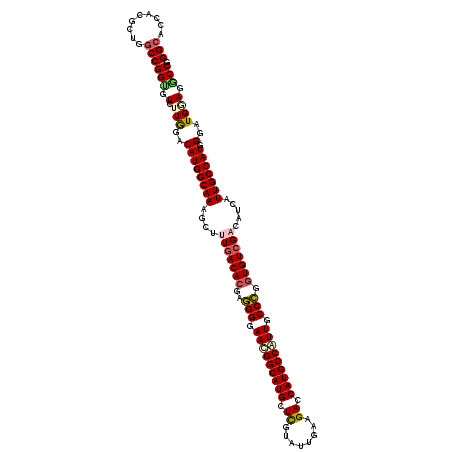
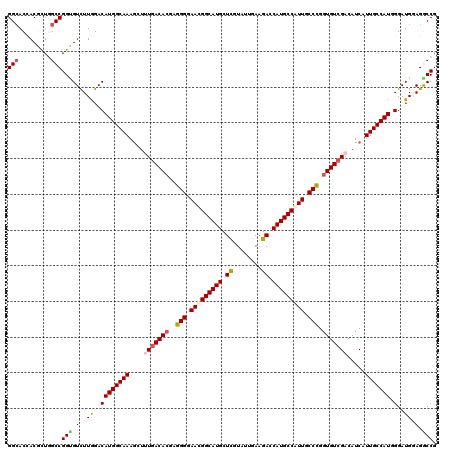
| Location | 10,643,369 – 10,643,489 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.78 |
| Mean single sequence MFE | -46.40 |
| Consensus MFE | -40.39 |
| Energy contribution | -40.87 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10643369 120 - 22224390 CGGUCUCCAUCCGAUGGCAAUGAUGUCGACACCGGGCAACGGCAUGGUCUUCAAUACGAGCAUGCCGUUCCCCUCGUGUCAAAGCUUUGCCAUGUCCAAGACUCCGGCUAGUGUGGUGCC .(((..((((((((((((((.(.....(((((.(((.(((((((((.((........)).))))))))).)))..)))))....).)))))))(((...)))..))).....)))).))) ( -48.70) >DroVir_CAF1 890 120 - 1 CGGUCUGCAUCCCAUGGCAAUGAUGUCGACACCGGGCAACGGCAUGGUCUUCAACACGAGCAUGCCGUUCCCCUCGUGUCAAAGCUUUGCCAUGUCCAAGACACCGGCCAGCGUGGUGCC ((((........((((((((.(.....(((((.(((.(((((((((.((........)).))))))))).)))..)))))....).))))))))........))))(((.....)))... ( -49.39) >DroPse_CAF1 718 120 - 1 CGGCCUUCAUCCCAUGGCAAUGAUGUCGACACCCGGCAAUGGCAUGGUCUUUAACACAAGCAUGCCAUUCCCAUCGUGUCAGAGCUUUGCCAUGUCCAAGACGCCGGCCAGCGUAGUUCC .((((..(.((.((((((((.(...(((((((..((.(((((((((..(((......)))))))))))).))...))))).)).).)))))))).....)).)..))))........... ( -40.50) >DroGri_CAF1 12163 120 - 1 CGGCCUGCAUCCAAUGGCAAUGAUGUCGACACCGGGCAAUGGCAUGGUCUUCAAUACGAGCAUGCCGUUCCCCUCCUGUCAAAGCUUUGCCAUGUCCAAAACACCGGCCAGCGUGGUGCC .((((..(.......((((....))))((((..(((.(((((((((.((........)).))))))))).)))...))))...(((..(((.(((.....)))..))).))))..).))) ( -44.00) >DroWil_CAF1 544 120 - 1 CGGCCUCCAUCCCAUGGCAAUGAUGUCUACACCGGGCAAUGGCAUGGUCUUUAAUACAAGCAUGCCGUUCCCCUCGUGUCAGGGCUUUGCCAUGUCUAAGACGCCGGCGAACGUUGUGCC .(((.((.....((((((((....((((((((.(((.(((((((((..(((......)))))))))))).)))..))))..)))).)))))))).....)).)))((((.......)))) ( -45.70) >DroAna_CAF1 691 120 - 1 CGGCCUCCAUCCAAUGGCAAUGAUGUCGACACCGGGCAACGGCAUGGUCUUCAAUACGAGCAUGCCGUUCCCCUCGUGUCAAAGUUUUGCCAUGUCCAAAACACCGGCCAGUGUGGUGCC .(((..((((....((((.........(((((.(((.(((((((((.((........)).))))))))).)))..)))))...((((((.......))))))....))))..)))).))) ( -50.10) >consensus CGGCCUCCAUCCCAUGGCAAUGAUGUCGACACCGGGCAACGGCAUGGUCUUCAAUACGAGCAUGCCGUUCCCCUCGUGUCAAAGCUUUGCCAUGUCCAAGACACCGGCCAGCGUGGUGCC .((((........(((((((.(.....(((((.(((.(((((((((.((........)).))))))))).)))..)))))....).)))))))(((...)))...))))........... (-40.39 = -40.87 + 0.48)


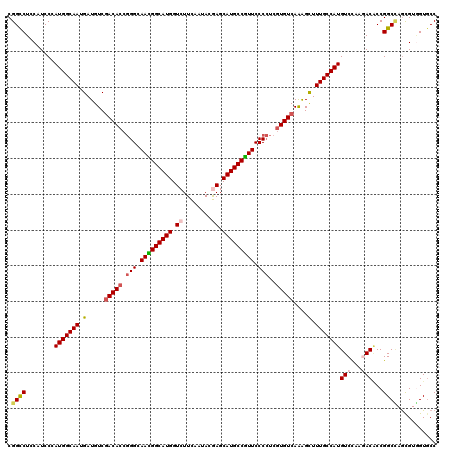
| Location | 10,643,409 – 10,643,529 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.22 |
| Mean single sequence MFE | -47.45 |
| Consensus MFE | -40.69 |
| Energy contribution | -40.87 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10643409 120 + 22224390 GACACGAGGGGAACGGCAUGCUCGUAUUGAAGACCAUGCCGUUGCCCGGUGUCGACAUCAUUGCCAUCGGAUGGAGACCGGGGAACAUGAGGCUCUGCGGCGCCAUCGCCUGCAGAUCGU (((((..(((.(((((((((.((........)).))))))))).))).)))))....((((..((.((((.......))))))...))))((.(((((((((....)))).))))))).. ( -54.70) >DroVir_CAF1 930 120 + 1 GACACGAGGGGAACGGCAUGCUCGUGUUGAAGACCAUGCCGUUGCCCGGUGUCGACAUCAUUGCCAUGGGAUGCAGACCGGGGAACAUAAGGCUCUGAGGCGCCAUUGCCUGCAGAUCGU (((((..(((.(((((((((.((........)).))))))))).))).)))))((.......(((.(((........)))(....)....)))(((((((((....))))).)))))).. ( -50.40) >DroPse_CAF1 758 120 + 1 GACACGAUGGGAAUGGCAUGCUUGUGUUAAAGACCAUGCCAUUGCCGGGUGUCGACAUCAUUGCCAUGGGAUGAAGGCCGGGGAACAUGAGGCUCUGCGGUGCCAUCGCCUGUAGGUCAU (((((..(((.((((((((((((......)))..))))))))).))).)))))(((.(((((.(...).))))).((((.(....)....))))((((((.((....))))))))))).. ( -47.70) >DroGri_CAF1 12203 120 + 1 GACAGGAGGGGAACGGCAUGCUCGUAUUGAAGACCAUGCCAUUGCCCGGUGUCGACAUCAUUGCCAUUGGAUGCAGGCCGGGGAACAUGAGGCUCUGUGGCGCAAUUGCCUGCAGAUCGU ....((.(((.((.((((((.((........)).)))))).)).)))((((....))))....))...((.(((((((((.(((.(.....).))).))))((....)))))))..)).. ( -41.90) >DroWil_CAF1 584 120 + 1 GACACGAGGGGAACGGCAUGCUUGUAUUAAAGACCAUGCCAUUGCCCGGUGUAGACAUCAUUGCCAUGGGAUGGAGGCCGGGGAACAUGAGGCUCUGUGGAGCCAUUGCCUGAAGAUCGU ...(((((((.((.(((((((((......)))..)))))).((.((((((.(...((((..........)))).).)))))).)).....(((((....))))).)).))).....)))) ( -40.60) >DroAna_CAF1 731 120 + 1 GACACGAGGGGAACGGCAUGCUCGUAUUGAAGACCAUGCCGUUGCCCGGUGUCGACAUCAUUGCCAUUGGAUGGAGGCCGGGGAACAUGAGACUCUGGGGUGCCAUCGCCUGCAGAUCAU (((((..(((.(((((((((.((........)).))))))))).))).)))))....((((..((.((((.......))))))...))))((.(((((((((....))))).)))))).. ( -49.40) >consensus GACACGAGGGGAACGGCAUGCUCGUAUUGAAGACCAUGCCAUUGCCCGGUGUCGACAUCAUUGCCAUGGGAUGGAGGCCGGGGAACAUGAGGCUCUGCGGCGCCAUCGCCUGCAGAUCGU (((((..(((.(((((((((.((........)).))))))))).))).)))))....((((..((...((.......))..))...))))((.(((((((.((....))))))))))).. (-40.69 = -40.87 + 0.17)

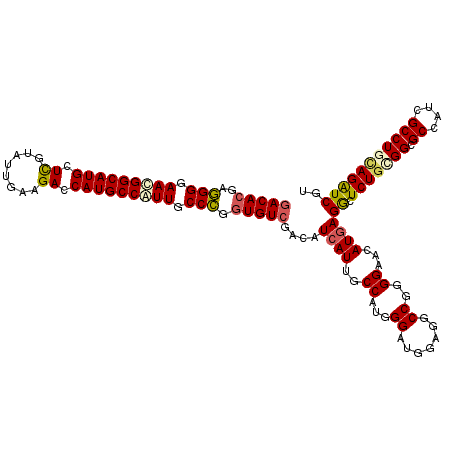

| Location | 10,643,409 – 10,643,529 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.22 |
| Mean single sequence MFE | -42.40 |
| Consensus MFE | -34.60 |
| Energy contribution | -34.72 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.571053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10643409 120 - 22224390 ACGAUCUGCAGGCGAUGGCGCCGCAGAGCCUCAUGUUCCCCGGUCUCCAUCCGAUGGCAAUGAUGUCGACACCGGGCAACGGCAUGGUCUUCAAUACGAGCAUGCCGUUCCCCUCGUGUC ....(((((.((((....)))))))))((.((((...((.(((.......)))..))..)))).)).(((((.(((.(((((((((.((........)).))))))))).)))..))))) ( -50.90) >DroVir_CAF1 930 120 - 1 ACGAUCUGCAGGCAAUGGCGCCUCAGAGCCUUAUGUUCCCCGGUCUGCAUCCCAUGGCAAUGAUGUCGACACCGGGCAACGGCAUGGUCUUCAACACGAGCAUGCCGUUCCCCUCGUGUC ..((((((.((((......))))))))(((..((((.(....)...)))).....))).......))(((((.(((.(((((((((.((........)).))))))))).)))..))))) ( -46.20) >DroPse_CAF1 758 120 - 1 AUGACCUACAGGCGAUGGCACCGCAGAGCCUCAUGUUCCCCGGCCUUCAUCCCAUGGCAAUGAUGUCGACACCCGGCAAUGGCAUGGUCUUUAACACAAGCAUGCCAUUCCCAUCGUGUC ..........((.(((((..(((..((((.....))))..)))...)))))))..((((....))))(((((..((.(((((((((..(((......)))))))))))).))...))))) ( -36.80) >DroGri_CAF1 12203 120 - 1 ACGAUCUGCAGGCAAUUGCGCCACAGAGCCUCAUGUUCCCCGGCCUGCAUCCAAUGGCAAUGAUGUCGACACCGGGCAAUGGCAUGGUCUUCAAUACGAGCAUGCCGUUCCCCUCCUGUC ......(((((((....))(((...((((.....))))...))))))))......((((....))))((((..(((.(((((((((.((........)).))))))))).)))...)))) ( -41.60) >DroWil_CAF1 584 120 - 1 ACGAUCUUCAGGCAAUGGCUCCACAGAGCCUCAUGUUCCCCGGCCUCCAUCCCAUGGCAAUGAUGUCUACACCGGGCAAUGGCAUGGUCUUUAAUACAAGCAUGCCGUUCCCCUCGUGUC .........(((((..(((((....)))))((((........(((..........))).)))))))))((((.(((.(((((((((..(((......)))))))))))).)))..)))). ( -37.10) >DroAna_CAF1 731 120 - 1 AUGAUCUGCAGGCGAUGGCACCCCAGAGUCUCAUGUUCCCCGGCCUCCAUCCAAUGGCAAUGAUGUCGACACCGGGCAACGGCAUGGUCUUCAAUACGAGCAUGCCGUUCCCCUCGUGUC ...(((((.((((..(((....)))..)))))).........(((..........)))...)))...(((((.(((.(((((((((.((........)).))))))))).)))..))))) ( -41.80) >consensus ACGAUCUGCAGGCAAUGGCGCCACAGAGCCUCAUGUUCCCCGGCCUCCAUCCCAUGGCAAUGAUGUCGACACCGGGCAACGGCAUGGUCUUCAAUACGAGCAUGCCGUUCCCCUCGUGUC ..(((....((((..((......))..))))...........(((..........)))......)))(((((.(((.(((((((((.((........)).))))))))).)))..))))) (-34.60 = -34.72 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:35 2006