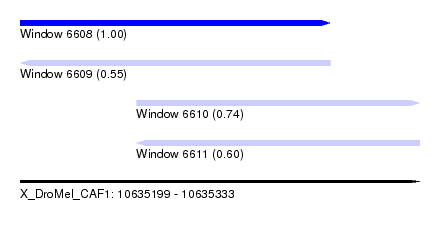
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,635,199 – 10,635,333 |
| Length | 134 |
| Max. P | 0.999332 |

| Location | 10,635,199 – 10,635,303 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -18.03 |
| Consensus MFE | -11.05 |
| Energy contribution | -12.55 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10635199 104 + 22224390 CAAAAUGAUAUACACAACAACAAAAUUAUA-AAAUAAUCAAAGUAAAU---AAGCGGGAAUCGU---AGAUUGAUUCCCA--------UAUGGGUAGUAAGAUAUCAACGGAAGAUAAA .....((((((..((.((............-...........((....---..))((((((((.---....)))))))).--------.....)).))...)))))).(....)..... ( -19.40) >DroEre_CAF1 17565 93 + 1 CAAAAUGAUAUACACAACAACAAAAUUAUA-AAAUAAUCAAAGUAAAU---AAGCGGGAAUCGC---AGAUUGAUUCCCA--------UAUGGGUAGUACGAUAUCAA----------- .....((((((..((.((............-...........((....---..))((((((((.---....)))))))).--------.....)).))...)))))).----------- ( -17.30) >DroYak_CAF1 17200 93 + 1 CAAAAUGAUAUACACAACAACAAAAUUAUA-AAAUAAUCAAAGUAAAU---AAGCGGGAAUCGC---AGAUUGAUUCCCA--------UAUGGGUUGUAAGAUAUCAA----------- .....((((((..(((((............-...........((....---..))((((((((.---....)))))))).--------.....)))))...)))))).----------- ( -21.40) >DroAna_CAF1 25114 105 + 1 CUAAAUGAAAUACACAACAAAAAAAUUAAACAAAUAAUCAAAGUAAAUAGUAAAGGGGAAUCAUCGGUGACGAAGUCCCAUCAUCUCCUAUGGAUA---CAAAAUCAA----------- ..........................................(((.((((.....((((....(((....)))..))))........))))...))---)........----------- ( -14.02) >consensus CAAAAUGAUAUACACAACAACAAAAUUAUA_AAAUAAUCAAAGUAAAU___AAGCGGGAAUCGC___AGAUUGAUUCCCA________UAUGGGUAGUAAGAUAUCAA___________ .....((((((.............(((((....))))).................((((((((........))))))))......................))))))............ (-11.05 = -12.55 + 1.50)

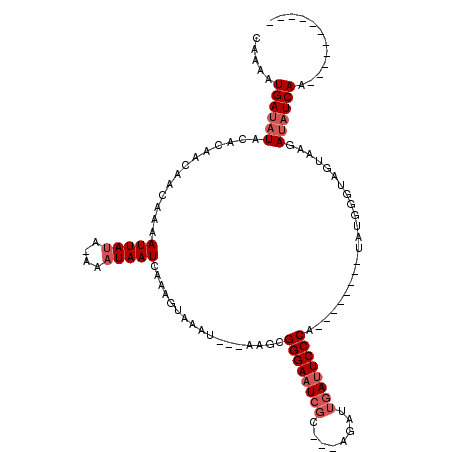
| Location | 10,635,199 – 10,635,303 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -16.79 |
| Consensus MFE | -11.19 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10635199 104 - 22224390 UUUAUCUUCCGUUGAUAUCUUACUACCCAUA--------UGGGAAUCAAUCU---ACGAUUCCCGCUU---AUUUACUUUGAUUAUUU-UAUAAUUUUGUUGUUGUGUAUAUCAUUUUG ............((((((..(((.((.....--------.(((((((.....---..)))))))....---....((...((((((..-.))))))..)).)).))).))))))..... ( -16.60) >DroEre_CAF1 17565 93 - 1 -----------UUGAUAUCGUACUACCCAUA--------UGGGAAUCAAUCU---GCGAUUCCCGCUU---AUUUACUUUGAUUAUUU-UAUAAUUUUGUUGUUGUGUAUAUCAUUUUG -----------.((((((.............--------.(((((((.....---..)))))))....---.................-((((((......)))))).))))))..... ( -15.60) >DroYak_CAF1 17200 93 - 1 -----------UUGAUAUCUUACAACCCAUA--------UGGGAAUCAAUCU---GCGAUUCCCGCUU---AUUUACUUUGAUUAUUU-UAUAAUUUUGUUGUUGUGUAUAUCAUUUUG -----------.((((((..((((((.....--------.(((((((.....---..)))))))....---....((...((((((..-.))))))..)).)))))).))))))..... ( -20.70) >DroAna_CAF1 25114 105 - 1 -----------UUGAUUUUG---UAUCCAUAGGAGAUGAUGGGACUUCGUCACCGAUGAUUCCCCUUUACUAUUUACUUUGAUUAUUUGUUUAAUUUUUUUGUUGUGUAUUUCAUUUAG -----------.........---..........((((((.((((..(((((...))))).))))...(((..........(((((......)))))..........)))..)))))).. ( -14.25) >consensus ___________UUGAUAUCGUACUACCCAUA________UGGGAAUCAAUCU___ACGAUUCCCGCUU___AUUUACUUUGAUUAUUU_UAUAAUUUUGUUGUUGUGUAUAUCAUUUUG ............((((((......................(((((((..........))))))).........................((((((......)))))).))))))..... (-11.19 = -12.00 + 0.81)


| Location | 10,635,238 – 10,635,333 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -14.29 |
| Energy contribution | -14.73 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10635238 95 + 22224390 AAGUAAAUAAGCGGGAAUCGUAGAUUGAUUCCCAUAUGGGUAGUAAGAUAUCAACGGAAGAUAAAACGGAAAUCAUUUUUCAAUAUGCCAAAUAA ............((((((((.....)))))))).....((((............(....).......(((((....)))))....))))...... ( -20.10) >DroEre_CAF1 17604 84 + 1 AAGUAAAUAAGCGGGAAUCGCAGAUUGAUUCCCAUAUGGGUAGUACGAUAUCAA-----------UCGGAAAUCAUUUUUCAAUAUGCCAAAUAA ............((((((((.....)))))))).....((((...((((....)-----------)))((((.....))))....))))...... ( -19.90) >DroYak_CAF1 17239 84 + 1 AAGUAAAUAAGCGGGAAUCGCAGAUUGAUUCCCAUAUGGGUUGUAAGAUAUCAA-----------UCGGAGAUAAUUUUUAAAUAUGCCAAAUAA ..........((((((((((.....)))))))).....(((((........)))-----------))...................))....... ( -16.40) >consensus AAGUAAAUAAGCGGGAAUCGCAGAUUGAUUCCCAUAUGGGUAGUAAGAUAUCAA___________UCGGAAAUCAUUUUUCAAUAUGCCAAAUAA ............((((((((.....)))))))).....((((....(....)...............(((((....)))))....))))...... (-14.29 = -14.73 + 0.45)


| Location | 10,635,238 – 10,635,333 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -15.70 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10635238 95 - 22224390 UUAUUUGGCAUAUUGAAAAAUGAUUUCCGUUUUAUCUUCCGUUGAUAUCUUACUACCCAUAUGGGAAUCAAUCUACGAUUCCCGCUUAUUUACUU ......(((((((..(.....(((.........))).....)..))))..............(((((((.......))))))))))......... ( -16.20) >DroEre_CAF1 17604 84 - 1 UUAUUUGGCAUAUUGAAAAAUGAUUUCCGA-----------UUGAUAUCGUACUACCCAUAUGGGAAUCAAUCUGCGAUUCCCGCUUAUUUACUU ......(((..........(((((.((...-----------..)).)))))...........(((((((.......))))))))))......... ( -16.30) >DroYak_CAF1 17239 84 - 1 UUAUUUGGCAUAUUUAAAAAUUAUCUCCGA-----------UUGAUAUCUUACAACCCAUAUGGGAAUCAAUCUGCGAUUCCCGCUUAUUUACUU ......(((............((((.....-----------..))))...............(((((((.......))))))))))......... ( -14.60) >consensus UUAUUUGGCAUAUUGAAAAAUGAUUUCCGA___________UUGAUAUCUUACUACCCAUAUGGGAAUCAAUCUGCGAUUCCCGCUUAUUUACUU ......(((.((((....))))........................................(((((((.......))))))))))......... (-12.40 = -12.73 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:27 2006