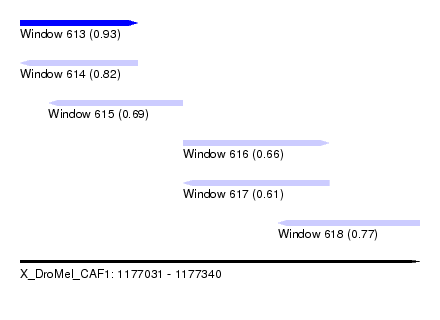
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,177,031 – 1,177,340 |
| Length | 309 |
| Max. P | 0.925209 |

| Location | 1,177,031 – 1,177,122 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -23.47 |
| Energy contribution | -23.29 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
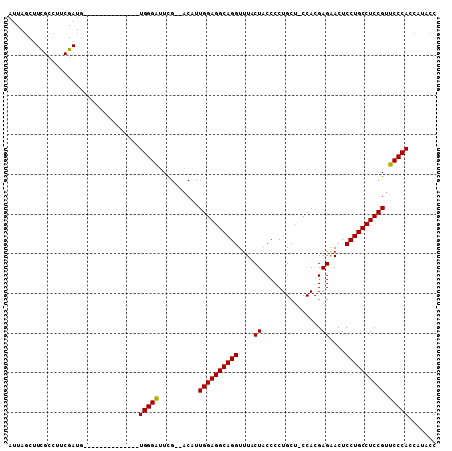
>X_DroMel_CAF1 1177031 91 + 22224390 AUUAGCUUCGCCUUCGAUG--------------UGGGAUUCG--ACAUUGGAGGCAGGUUUACUGCCCCUGCU-CCACGAGAACUCCUGCCUCCGUUCCCAGCAGACC .........((((((((((--------------(.(....).--))))))))))).(((.....))).(((((-..(((.((.........)))))....)))))... ( -27.60) >DroSec_CAF1 12994 90 + 1 AUUAGCUUUGCCUUCGAUG--------------UGGGAUUCG--AC-UUGGAGGCAGGUUUACUACCCCUGCU-CCACGAGAACUCCUGCCUCCGUUCCCACCAUACC ..................(--------------(((((....--..-.((((((((((.............((-(...)))....)))))))))).))))))...... ( -26.73) >DroSim_CAF1 28059 92 + 1 AUUAGCUUCGCCUUCGAUG--------------UGGGAUUCG--ACAUUGGAGGCAGGUUUACUACCCCUGCUUCCACGAGAACUCCUGCCUCCGUUCCCACCAUACC ..................(--------------(((((..((--....((((((((((.........)))))))))).(((.........))))).))))))...... ( -28.20) >DroEre_CAF1 12247 102 + 1 AUUAACUUCGCCUUCGAUUCUCGACAUUGGCUUUGGGGUUUGCCCCUUUGGAGGCAGGUUUACUGCCCCUGCG-CCACGAGGACUCCUGCCUCCGUUCCCAC-----C .........(((.(((.....)))....)))...((((....))))...(((((((((......((....)).-((....))...)))))))))........-----. ( -34.50) >consensus AUUAGCUUCGCCUUCGAUG______________UGGGAUUCG__ACAUUGGAGGCAGGUUUACUACCCCUGCU_CCACGAGAACUCCUGCCUCCGUUCCCACCAUACC .................................(((((..........((((((((((....((...............))....)))))))))).)))))....... (-23.47 = -23.29 + -0.19)

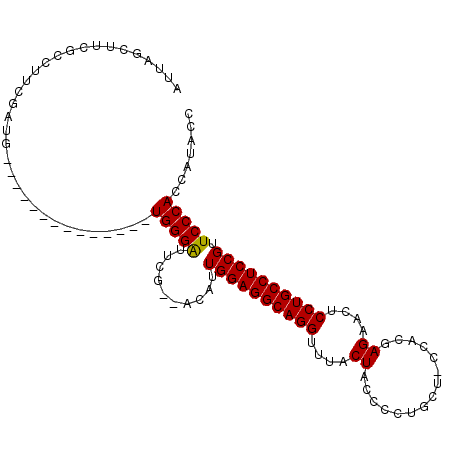
| Location | 1,177,031 – 1,177,122 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177031 91 - 22224390 GGUCUGCUGGGAACGGAGGCAGGAGUUCUCGUGG-AGCAGGGGCAGUAAACCUGCCUCCAAUGU--CGAAUCCCA--------------CAUCGAAGGCGAAGCUAAU .((((.(((....)))))))...((((.((((((-((((((.........)))).))))....(--(((......--------------..))))..))))))))... ( -29.40) >DroSec_CAF1 12994 90 - 1 GGUAUGGUGGGAACGGAGGCAGGAGUUCUCGUGG-AGCAGGGGUAGUAAACCUGCCUCCAA-GU--CGAAUCCCA--------------CAUCGAAGGCAAAGCUAAU ......(((((((((((((((((....(((.((.-..)).))).......)))))))))..-))--....)))))--------------)......(((...)))... ( -29.70) >DroSim_CAF1 28059 92 - 1 GGUAUGGUGGGAACGGAGGCAGGAGUUCUCGUGGAAGCAGGGGUAGUAAACCUGCCUCCAAUGU--CGAAUCCCA--------------CAUCGAAGGCGAAGCUAAU (((...(((((((((((((((((....(((.((....)).))).......)))))))))...))--....)))))--------------).(((....))).)))... ( -33.80) >DroEre_CAF1 12247 102 - 1 G-----GUGGGAACGGAGGCAGGAGUCCUCGUGG-CGCAGGGGCAGUAAACCUGCCUCCAAAGGGGCAAACCCCAAAGCCAAUGUCGAGAAUCGAAGGCGAAGUUAAU .-----.....((((((((((((.((((..(...-..)..))))......)))))))))...((((....))))...(((....(((.....))).)))...)))... ( -39.20) >consensus GGUAUGGUGGGAACGGAGGCAGGAGUUCUCGUGG_AGCAGGGGCAGUAAACCUGCCUCCAAUGU__CGAAUCCCA______________CAUCGAAGGCGAAGCUAAU .......(((((..(((((((((....(((.((....)).))).......)))))))))...(...)...)))))................................. (-25.02 = -25.28 + 0.25)

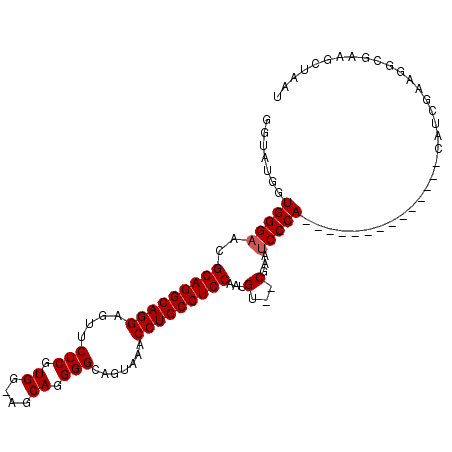
| Location | 1,177,053 – 1,177,157 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -45.66 |
| Consensus MFE | -33.45 |
| Energy contribution | -34.70 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177053 104 - 22224390 -----UCAUGCCCAGCGCGUGCGUGGCGUUACGCUGGGCUGGUCUGCUGGGAACGGAGGCAGGAGUUCUCGUGG-AGCAGGGGCAGUAAACCUGCCUCCAAUGU--CGAAUC-------- -----....((((((((...(((...)))..))))))))((.((..(.((((((..........)))))))..)-).))(((((((.....))))).)).....--......-------- ( -42.80) >DroSec_CAF1 13016 106 - 1 C--CUUCAUGUCCAGCGCGUGCGUGGCGUUACGCUGUGCUGGUAUGGUGGGAACGGAGGCAGGAGUUCUCGUGG-AGCAGGGGUAGUAAACCUGCCUCCAA-GU--CGAAUC-------- (--(.(((((.((((((((.(((((....)))))))))))))))))).))((..(((((((((....(((.((.-..)).))).......)))))))))..-.)--).....-------- ( -47.60) >DroSim_CAF1 28081 108 - 1 C--CUUCAUGUCCAGCGCGUGCGUGGCGUUACGCUGUGCUGGUAUGGUGGGAACGGAGGCAGGAGUUCUCGUGGAAGCAGGGGUAGUAAACCUGCCUCCAAUGU--CGAAUC-------- (--(.(((((.((((((((.(((((....)))))))))))))))))).))....(((((((((....(((.((....)).))).......))))))))).....--......-------- ( -50.40) >DroEre_CAF1 12283 92 - 1 --------------GCGCGUGCGUGGCGUUCCGCUCUGCUG-----GUGGGAACGGAGGCAGGAGUCCUCGUGG-CGCAGGGGCAGUAAACCUGCCUCCAAAGGGGCAAACC-------- --------------((.(.(((((.(((....((((((((.-----.((....))..)))).))))...))).)-)))).).))........((((((....))))))....-------- ( -40.60) >DroYak_CAF1 15126 114 - 1 CAUCUUCAUGCCCAGCGCGUGCGUGGCGUUCCGCUGUGGUG-----AAGGGAACGGAGGCAGGAGUCCUCGUGG-CGCAGGGGCAGUAAACCUGCCUCCAGAGGGGCAAACCAAAGGGGC .........((((.((.(.(((((.(((((((.((......-----.))))))).(((((....).)))))).)-)))).).))........((((((....))))))........)))) ( -46.90) >consensus C__CUUCAUGCCCAGCGCGUGCGUGGCGUUACGCUGUGCUGGU_UGGUGGGAACGGAGGCAGGAGUUCUCGUGG_AGCAGGGGCAGUAAACCUGCCUCCAAAGU__CGAAUC________ ...........((((((((.(((........)))))))))))......(....)(((((((((....(((.((....)).))).......)))))))))..................... (-33.45 = -34.70 + 1.25)


| Location | 1,177,157 – 1,177,270 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -28.02 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177157 113 + 22224390 -AGAUCGGGAGACGC------AGAUCGAAGAGGACGCAGAGGAAUUUUGUGCGCAGGCGCAGCCCAAAAAGGGCACCAACCUCUUCAAGAGAUCUGGCUUCCCAGCUUUGCUGCCUGUUU -.....((((..(.(------((((((((((((..............(((((....)))))((((.....)))).....)))))))....)))))))..)))).((......))...... ( -41.50) >DroSec_CAF1 13122 112 + 1 AAGAUCGGGAGACGC------AGAUCGAAGAGGACGCAGAGGAAUUUUGUGCGCAGGCGCAGCCCAAAAAGGGCACCAACCUCUUUAA--GAUCUGGCUUCCCAGCUUUGCGGCCUGUUU .((..(((((..(.(------((((((((((((..............(((((....)))))((((.....)))).....)))))))..--)))))))..)))).((...)))..)).... ( -40.90) >DroSim_CAF1 28189 112 + 1 AAGAUCGGGAGACGC------AGAUCGAAGAGGACGCAGAGGAAUUUUGUGCGCAGGCGCAGCCCAAAAAGGGCACCAACCUCUUUAA--GAUCUGGCUUCCCAGCUUUGCGGCCUGUUU .((..(((((..(.(------((((((((((((..............(((((....)))))((((.....)))).....)))))))..--)))))))..)))).((...)))..)).... ( -40.90) >DroEre_CAF1 12375 97 + 1 ---------------------GGAUCGAAGAGGACGCCGAGGAAUUUUAUGCGCAGGCGCAGCCCAAAAAGGGCACCAACCUCUCCAA--GAUCUUGCUUCCCAGCUUUGCUGCCUGCUU ---------------------(((((..(((((.((((..(.(......).)...))))..((((.....)))).....)))))....--))))).((....((((...))))...)).. ( -30.00) >DroYak_CAF1 15240 118 + 1 AAGAUGGGAAGAUGGGAAGGAGAAUCGGAGAGGACGCGGAGGAAUUUUAUGCGCAGGCGCAGCCCAAAAGGGGCAGCAACCUCUUCAU--GAUCUUGCUUCCCAGCUUUGCUGCCUGCUU .....(.((((.(((((((.(..((((((((((..((............((((....))))((((.....)))).))..)))))))..--)))..).))))))).)))).)......... ( -46.10) >consensus AAGAUCGGGAGACGC______AGAUCGAAGAGGACGCAGAGGAAUUUUGUGCGCAGGCGCAGCCCAAAAAGGGCACCAACCUCUUCAA__GAUCUGGCUUCCCAGCUUUGCUGCCUGUUU ......(((((..........((((((((((((................((((....))))((((.....)))).....)))))))....)))))..)))))((((......).)))... (-28.02 = -28.70 + 0.68)

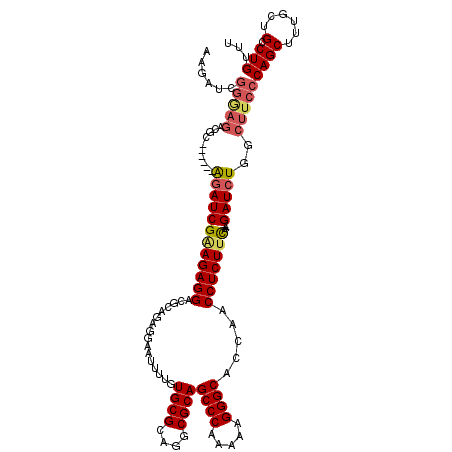
| Location | 1,177,157 – 1,177,270 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177157 113 - 22224390 AAACAGGCAGCAAAGCUGGGAAGCCAGAUCUCUUGAAGAGGUUGGUGCCCUUUUUGGGCUGCGCCUGCGCACAAAAUUCCUCUGCGUCCUCUUCGAUCU------GCGUCUCCCGAUCU- .....(((......)))((((..(((((((....(((((((.((..((((.....))))((((....)))).............)).))))))))))))------).)..)))).....- ( -42.80) >DroSec_CAF1 13122 112 - 1 AAACAGGCCGCAAAGCUGGGAAGCCAGAUC--UUAAAGAGGUUGGUGCCCUUUUUGGGCUGCGCCUGCGCACAAAAUUCCUCUGCGUCCUCUUCGAUCU------GCGUCUCCCGAUCUU .....(((......)))((((..(((((((--...((((((.((..((((.....))))((((....)))).............)).)))))).)))))------).)..))))...... ( -40.10) >DroSim_CAF1 28189 112 - 1 AAACAGGCCGCAAAGCUGGGAAGCCAGAUC--UUAAAGAGGUUGGUGCCCUUUUUGGGCUGCGCCUGCGCACAAAAUUCCUCUGCGUCCUCUUCGAUCU------GCGUCUCCCGAUCUU .....(((......)))((((..(((((((--...((((((.((..((((.....))))((((....)))).............)).)))))).)))))------).)..))))...... ( -40.10) >DroEre_CAF1 12375 97 - 1 AAGCAGGCAGCAAAGCUGGGAAGCAAGAUC--UUGGAGAGGUUGGUGCCCUUUUUGGGCUGCGCCUGCGCAUAAAAUUCCUCGGCGUCCUCUUCGAUCC--------------------- ..((...((((...))))....))..((((--..(((((((.((.(((((.....))))((((....))))...........).)).))))))))))).--------------------- ( -31.00) >DroYak_CAF1 15240 118 - 1 AAGCAGGCAGCAAAGCUGGGAAGCAAGAUC--AUGAAGAGGUUGCUGCCCCUUUUGGGCUGCGCCUGCGCAUAAAAUUCCUCCGCGUCCUCUCCGAUUCUCCUUCCCAUCUUCCCAUCUU ..((.....)).(((.(((((((.(..(((--..((.((((.(((.((((.....))))((((....))))............))).)))))).)))..).))))))).)))........ ( -37.90) >consensus AAACAGGCAGCAAAGCUGGGAAGCCAGAUC__UUGAAGAGGUUGGUGCCCUUUUUGGGCUGCGCCUGCGCACAAAAUUCCUCUGCGUCCUCUUCGAUCU______GCGUCUCCCGAUCUU ..............(((....)))..((((....(((((((.((..((((.....))))((((....)))).............)).)))))))))))...................... (-25.94 = -26.26 + 0.32)

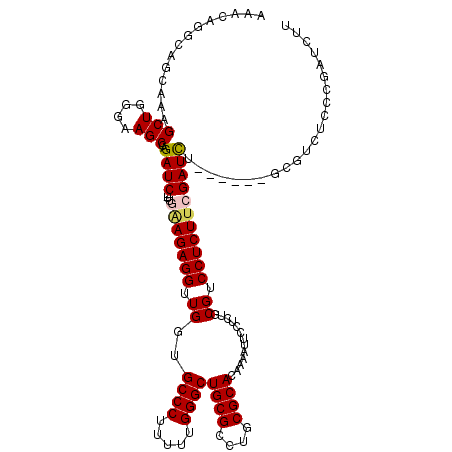
| Location | 1,177,230 – 1,177,340 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.46 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -18.19 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1177230 110 - 22224390 GAGAUGCUGCCGCCAGCUGG---CUU-------UCGGUGUGGACUGUCAGCAGCUUACCUAAACAAAAAAAAAAAAAAAAAAACAGGCAGCAAAGCUGGGAAGCCAGAUCUCUUGAAGAG ((((((((((((((....))---)..-------..((((.((.(((....)))))))))..........................))))))....((((....)))))))))........ ( -36.20) >DroSec_CAF1 13196 102 - 1 GAGACGCUGCCGCCAGCUGG---CUG-------UCGGUGUGGAUUGUCAGCAGCCUACCUAAACAAAAAA------AAAAAAACAGGCCGCAAAGCUGGGAAGCCAGAUC--UUAAAGAG ((((.((.((((((....))---)..-------..((((.((.(((....)))))))))...........------.........))).))....((((....)))).))--))...... ( -28.80) >DroSim_CAF1 28263 100 - 1 GAGACGCUGCCGCCAGCUGG---CUG-------UCGGUGUGGAUUGUCAGCAGCCUUCCUAAACAAAAA--------AAAAAACAGGCCGCAAAGCUGGGAAGCCAGAUC--UUAAAGAG ((((...(((.(((.(((((---(.(-------((......))).))))))((.....)).........--------........))).)))...((((....)))).))--))...... ( -27.40) >DroEre_CAF1 12434 100 - 1 GAGAUGCUGCCGCCAGCAGCUGGCUGCCGGCAGUUGGUGCGGAUUGUCGG---CCUACCCAA---------------AAAAAGCAGGCAGCAAAGCUGGGAAGCAAGAUC--UUGGAGAG ((((((((((((((((...))))).((((((((((......)))))))))---)........---------------........))))))...(((....)))...)))--))...... ( -40.50) >consensus GAGACGCUGCCGCCAGCUGG___CUG_______UCGGUGUGGAUUGUCAGCAGCCUACCUAAACAAAAA________AAAAAACAGGCAGCAAAGCUGGGAAGCCAGAUC__UUAAAGAG .....((((((..(((.......))).........((((.((.(((....)))))))))..........................))))))....((((....))))............. (-18.19 = -19.88 + 1.69)

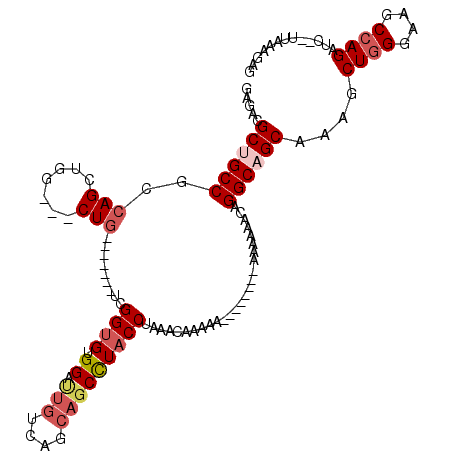
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:44 2006