
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,633,164 – 10,633,271 |
| Length | 107 |
| Max. P | 0.546338 |

| Location | 10,633,164 – 10,633,271 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -20.87 |
| Energy contribution | -21.87 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
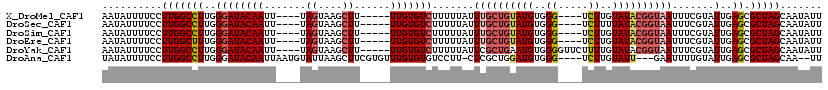
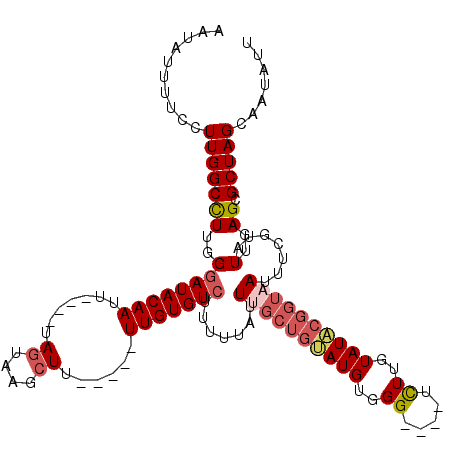
>X_DroMel_CAF1 10633164 107 + 22224390 AAUAUUUUCCUUGGCCUUGGGAUACAAUU----UAGUAAGCUU-----UUGUGUCUUUUUAUUUGCUGUAUGUGGG----UCUUGUAUACGGUAAUUUCGUAUUGAGCGCUAGCAAUAUU ..........(((((((..((((((((..----.((....)).-----))))))).......((((((((((..(.----..)..)))))))))).......)..)).)))))....... ( -25.00) >DroSec_CAF1 14920 107 + 1 AAUAUUUUCCUUGGCCUUGGGAUACAAUU----UAGUAAGCUU-----UUGUGUCUUUUUAUUUGCUGUAUGUGGG----UCUUUUAUACGGUAAUUUCGUAUUGAGCGCUAGCAAUAUU ..........(((((((..((((((((..----.((....)).-----))))))).......((((((((((....----.....)))))))))).......)..)).)))))....... ( -23.40) >DroSim_CAF1 15486 107 + 1 AAUAUUUUCCUUGGCCUUGGGAUACAAUU----UAGUAAGCUU-----UUGUGUCUUUUUAUUUGCUGUAUGUGGG----UCUUGUAUACGGUAAUUUCGUAUUGAGCGCUAGCAAUAUU ..........(((((((..((((((((..----.((....)).-----))))))).......((((((((((..(.----..)..)))))))))).......)..)).)))))....... ( -25.00) >DroEre_CAF1 15570 107 + 1 AAUAUUUUCCUUGGCUUUGGGAUACAAUU----UAGUAAGCUU-----UUGUGUCUUUUUAUUUGCUGUAUGUGGG----UCUUGUAUACGGUAAUUUCGUAUUGAGCGCUAGCAAUAUU ..........(((((...(((((((((..----.((....)).-----))))))))).....((((((((((..(.----..)..)))))))))).............)))))....... ( -22.60) >DroYak_CAF1 15147 111 + 1 AAUAUUUUCCUUGGCCUUGGGAUACAAUU----UAGUAAGCUU-----UUGUGUCUUUUUAUUCGCUGAAUGUGGGGUUCUUUUGUAUACGGUAAUUUCGUAUUGAGCGCUAGCAAUAUU ..........(((((...(((((((((..----.((....)).-----))))))))).......(((.((((((..(((...(((....))).)))..)))))).))))))))....... ( -23.70) >DroAna_CAF1 23055 110 + 1 UAUAUUUUCCUUGGCCUUGGGAUACAAUUAAUGUAUUAAGCUUCGUGUUUGUGUGUCCUU-CUCGCUGGAUGUGGG----UCUUGUAUU---GAAUUUUGUAUUGAGCGCUAGCAA--UU ..........(((((...((((((((....(((..........))).....)))))))).-...(((.((((..(.----((.......---))...)..)))).))))))))...--.. ( -23.00) >consensus AAUAUUUUCCUUGGCCUUGGGAUACAAUU____UAGUAAGCUU_____UUGUGUCUUUUUAUUUGCUGUAUGUGGG____UCUUGUAUACGGUAAUUUCGUAUUGAGCGCUAGCAAUAUU ..........(((((((..((((((((.......((....))......))))))).......((((((((((..((.....))..)))))))))).......)..)).)))))....... (-20.87 = -21.87 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:21 2006