| Sequence ID | X_DroMel_CAF1 |
|---|---|
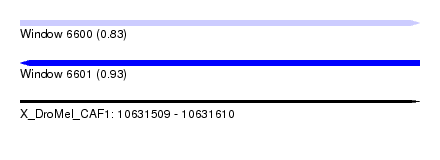
| Location | 10,631,509 – 10,631,610 |
| Length | 101 |
| Max. P | 0.925260 |

| Location | 10,631,509 – 10,631,610 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10631509 101 + 22224390 GGCCUGCCAACUCGUUGCUA-UUGCAUAAGUUGGCAGUUCCAGCGUGGUUGCA-------A---AUUGGAAGGAAAUAGAUGG--GAAAAAGAUCCAGCGUGGCUUCU-UCCAUU ((.(((((((((...(((..-..)))..)))))))))..)).(((....))).-------.---..((((((((..(((...(--((......)))..).))..))))-)))).. ( -34.40) >DroPse_CAF1 14169 111 + 1 GCACUGCCAACUCACCGA-AAUUGCAUAAGUUGGCAGUUCCAGCGUGGUUGCAAAAAAAAAAAAACUAAAACCAAAUGUAUG--AAAAAAUGAUCCAGCGUGGCUGGAAUA-GUU ..((((((((((......-.........))))))))))((((((.(((((...................))))).((((..(--(........))..)))).))))))...-... ( -27.87) >DroSec_CAF1 13093 110 + 1 GGCCUGCCAACUCGUUGCUAAUUGCAUAAGUUGGCAGUUCCAGCGUGGUUGCAAAAAAAAA---AUUGGAAGGAAAAAGAUGG-GGAAAAAGAUCCAGCGUGGCUUCU-UCCAUU ((.(((((((((...(((.....)))..)))))))))..)).(((....))).........---..((((((((......(((-(........)))).......))))-)))).. ( -34.62) >DroSim_CAF1 13191 110 + 1 GGCCUGCCAACUCGUUGCUAAUUGCAUAAGUUGGCAGUUCCAGCGUGGUUGCAAAAAAAAA---AUUGGAAGGAAAAAGAUGG-GGAAAAAGAUCCAGCGUGGCUUCU-UCCAUU ((.(((((((((...(((.....)))..)))))))))..)).(((....))).........---..((((((((......(((-(........)))).......))))-)))).. ( -34.62) >DroEre_CAF1 12978 111 + 1 GGCCUGCCAACUCGUUGCCAAUUGCAUAAGUUGGCAGUUCCAGCGUGGUUGCAAAAAAAAA---A-UGAAGGAAAAAAGAUGGGGGAAAAUGAUCCAGCGUGGCUUCUAUCAAUU ...(((((((((...(((.....)))..)))))))))((((..(((..((.......))..---)-))..))))....((((((((...(((......)))..)))))))).... ( -32.70) >DroYak_CAF1 13838 97 + 1 GGCCUGCCAACUCGUUGCCAGUUGCGUAAGUUGGCAGUUCCAGCGUGGUUGCAAAAAAAAA---A---------------UGGGAGAAAAUGAUCCAGCGUGGCUUCUAUAAAUU ((.(((((((((...(((.....)))..)))))))))..)).(((....))).........---(---------------((((((...(((......)))..)))))))..... ( -27.70) >consensus GGCCUGCCAACUCGUUGCUAAUUGCAUAAGUUGGCAGUUCCAGCGUGGUUGCAAAAAAAAA___AUUGGAAGGAAAAAGAUGG_GGAAAAAGAUCCAGCGUGGCUUCU_UCCAUU ...(((((((((...(((.....)))..)))))))))..(((((.(((..............................................))))).)))............ (-20.54 = -20.77 + 0.22)

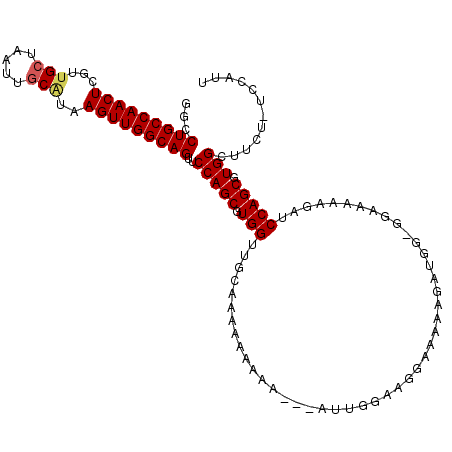

| Location | 10,631,509 – 10,631,610 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -21.79 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10631509 101 - 22224390 AAUGGA-AGAAGCCACGCUGGAUCUUUUUC--CCAUCUAUUUCCUUCCAAU---U-------UGCAACCACGCUGGAACUGCCAACUUAUGCAA-UAGCAACGAGUUGGCAGGCC ...(((-(((((((.....))..)))))))--)...........(((((..---.-------.((......)))))))((((((((((.(((..-..)))..))))))))))... ( -32.20) >DroPse_CAF1 14169 111 - 1 AAC-UAUUCCAGCCACGCUGGAUCAUUUUUU--CAUACAUUUGGUUUUAGUUUUUUUUUUUUUGCAACCACGCUGGAACUGCCAACUUAUGCAAUU-UCGGUGAGUUGGCAGUGC ...-...((((((.....((((.......))--))......(((((.(((...........))).))))).))))))(((((((((((((......-...))))))))))))).. ( -32.20) >DroSec_CAF1 13093 110 - 1 AAUGGA-AGAAGCCACGCUGGAUCUUUUUCC-CCAUCUUUUUCCUUCCAAU---UUUUUUUUUGCAACCACGCUGGAACUGCCAACUUAUGCAAUUAGCAACGAGUUGGCAGGCC ..((((-((.(((...)))(((......)))-...........))))))..---.........((..((.....))..((((((((((.(((.....)))..)))))))))))). ( -32.10) >DroSim_CAF1 13191 110 - 1 AAUGGA-AGAAGCCACGCUGGAUCUUUUUCC-CCAUCUUUUUCCUUCCAAU---UUUUUUUUUGCAACCACGCUGGAACUGCCAACUUAUGCAAUUAGCAACGAGUUGGCAGGCC ..((((-((.(((...)))(((......)))-...........))))))..---.........((..((.....))..((((((((((.(((.....)))..)))))))))))). ( -32.10) >DroEre_CAF1 12978 111 - 1 AAUUGAUAGAAGCCACGCUGGAUCAUUUUCCCCCAUCUUUUUUCCUUCA-U---UUUUUUUUUGCAACCACGCUGGAACUGCCAACUUAUGCAAUUGGCAACGAGUUGGCAGGCC ...(((..(((((...)))(((......)))...........))..)))-.---.........((..((.....))..((((((((((.(((.....)))..)))))))))))). ( -26.10) >DroYak_CAF1 13838 97 - 1 AAUUUAUAGAAGCCACGCUGGAUCAUUUUCUCCCA---------------U---UUUUUUUUUGCAACCACGCUGGAACUGCCAACUUACGCAACUGGCAACGAGUUGGCAGGCC ............(((.(((((..((..........---------------.---........))...))).)))))..((((((((((..((.....))...))))))))))... ( -23.31) >consensus AAUGGA_AGAAGCCACGCUGGAUCAUUUUCC_CCAUCUUUUUCCUUCCAAU___UUUUUUUUUGCAACCACGCUGGAACUGCCAACUUAUGCAAUUAGCAACGAGUUGGCAGGCC ............(((.(((((..............................................))).)))))..((((((((((.(((.....)))..))))))))))... (-21.79 = -22.02 + 0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:18 2006