| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,631,119 – 10,631,216 |
| Length | 97 |
| Max. P | 0.763564 |

| Location | 10,631,119 – 10,631,216 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 67.46 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -4.43 |
| Energy contribution | -6.07 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.17 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10631119 97 - 22224390 GAAGAAGAGAGAGAG--AGUUGGUUUUA--UUUCUUAAGAGU-AAU-GUAACUAUUUCUCUUUGGUUGC----------CUCCCUUUCUUUUCUGACCUGCUUUCUUUUAUUA ..(((((((((((.(--(((((.(((((--.....))))).)-)).-(((((((........)))))))----------))).)))))))))))................... ( -25.00) >DroPse_CAF1 13793 96 - 1 GAC--------------UG-UGCUUCUGUAUUUGU-AAAAGAACAU-GUAACUGUGUUGCUUUUGUCGCUUGUGAUUUCUUUCCCUUCUUCUUUGACCCGCUUUCUUCGGGAA (((--------------.(-(((....))))....-((((((((((-......))))).)))))))).............................((((.......)))).. ( -14.90) >DroSec_CAF1 12711 100 - 1 GAGGAAGAGAGAGAGACAGUUGCUUCUA--UUUCUUAAGAGU-AAU-GUAAUCAUUUCUCUUUGGUUGC---------CCUCCCUUUCUCUUCUGACCUGCUUUCUUUUAUAA ..(((((((((((.((..((((((..((--.....))..)))-)))-(((((((........)))))))---------..)).)))))))))))................... ( -27.60) >DroSim_CAF1 12807 100 - 1 GAGGAAGAGAGAGAGACAGUUGCUUCUA--UUUCUUAAGAGU-AGU-GUAACCAUUUCUCUUUGGUUGC---------CCUCCCUUUCUCUUCUGACCUGCUUUCUUUUAUAA ..(((((((((((.((...(((((..((--.....))..)))-)).-(((((((........)))))))---------..)).)))))))))))................... ( -28.30) >DroYak_CAF1 13460 96 - 1 GAGGAAGAGA----GAGAGUUGGUUUUA--UAUCUUAAGAGU-AAU-GUAACUAUCUCUCUUUGGUCGC---------CCUCCCUUUCUCUUCUGACCUGUUUUCUUUUAUUA ..((((((((----(((.(..(((....--.(((..(((((.-.((-(....)))..))))).))).))---------)..).)))))))))))................... ( -23.30) >DroAna_CAF1 20948 85 - 1 AAAGAGGAGAGAGCGGGAGG---------------UCAGAGA-AAUGGUAACUAUUGCUCUUAGGAG------------CCGCGUUUCUCUUCUGACCCGCUUUCUUUUGGCG ....(((((((((((((.((---------------..(((((-((((((..(((.......)))..)------------))..)))))))).))..))))))))))))).... ( -34.90) >consensus GAGGAAGAGAGAGAGA_AGUUGCUUCUA__UUUCUUAAGAGU_AAU_GUAACUAUUUCUCUUUGGUUGC_________CCUCCCUUUCUCUUCUGACCUGCUUUCUUUUAUAA ..(((((((((.........................(((((................))))).((((...........................))))..))))))))).... ( -4.43 = -6.07 + 1.64)


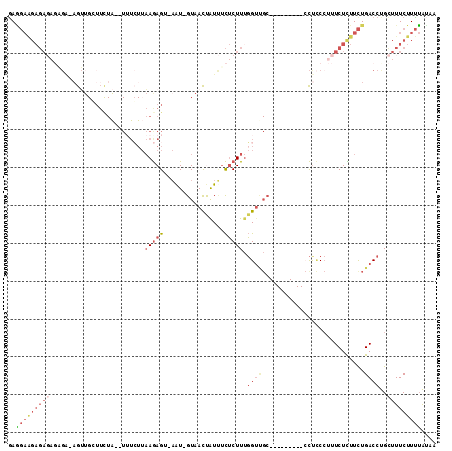
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:16 2006