| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,609,628 – 10,609,726 |
| Length | 98 |
| Max. P | 0.606549 |

| Location | 10,609,628 – 10,609,726 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.42 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.56 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
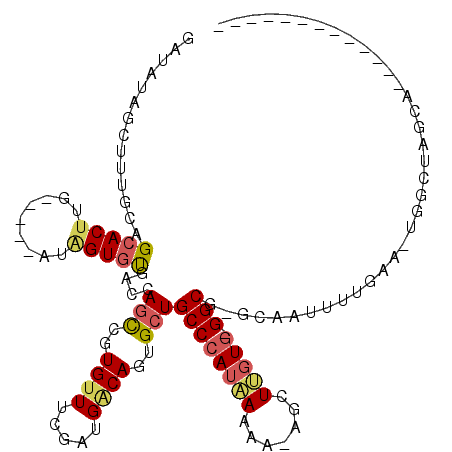
>X_DroMel_CAF1 10609628 98 + 22224390 GAUAUUGCUUUGCAGCACUUG-----AUGGUGUGACCAGCUGUGUUUUGAUGACAGUGCUGCCCAUAAAAA-CGUUUGUG-GGCG-GCAAUUUUGAA-UGGCUAGCA------------- .....((((..(((((((...-----.(((.....)))...)))))..........(((((((((((((..-..))))))-))))-)))........-..)).))))------------- ( -33.70) >DroSec_CAF1 6770 99 + 1 GAUAUUGCUUUGCUGAACUUG-----AAAGUGUGACCAGCAAUGUUUCGAUGGCAGUGCUGCCCAUAAAAA-CGUUUGUG-GGCG-GCAAUUUUGAAUUGGCUAGCA------------- .....((((..((..(..(..-----(((...((.((((((.((((.....)))).))))(((((((((..-..))))))-))))-))).)))..).)..)).))))------------- ( -33.10) >DroEre_CAF1 6742 97 + 1 GAUAUAGCUUUGCAGCACUUG-----AUAGUGCGACCAGCCGUGUUUCGGUGGCAGUGCUGCCCAUAAUUAAAGCUUGUG-GGCG-CUAGUUUUGAA-UGG--AACA------------- ....((((...((((((((..-----..)))))(.(((.(((.....)))))))..))).((((((((.......)))))-))))-)))(((((...-.))--))).------------- ( -34.20) >DroYak_CAF1 7331 97 + 1 GAU-UAGCUUUGCAGCACACG-----AUAGUGUGACCAGCCGUGUUUCGGUGACAGUGCUGCCCAUUAAUG-AGCUUUUG-GGCG-CCAGUUUUGAA-UGACAGACA------------- ...-.(((.((((..(((((.-----...)))))....((((.....))))).))).)))(((((......-......))-))).-...(((.....-.))).....------------- ( -24.30) >DroAna_CAF1 9050 109 + 1 -------CUGUCCAGCACUUGGCCAAAAAGUGUGGCCAAUUUUGUUAAG--GCCAGUGCUGCCCAUCAAGU-GGAUGGUAAGGCGCGCAAUUCAAAA-UAGCUAGACUUGCUACUUAGUG -------..(((.(((...((((((.......))))))((((((.....--((..(((((..(((((....-.)))))...)))))))....)))))-).))).)))............. ( -34.70) >consensus GAUAUAGCUUUGCAGCACUUG_____AUAGUGUGACCAGCCGUGUUUCGAUGACAGUGCUGCCCAUAAAAA_AGCUUGUG_GGCG_GCAAUUUUGAA_UGGCUAGCA_____________ ..............(((((.........)))))....(((..((((.....))))..)))((((((((.......))))).))).................................... (-11.00 = -11.56 + 0.56)

| Location | 10,609,628 – 10,609,726 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.42 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -8.09 |
| Energy contribution | -8.89 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
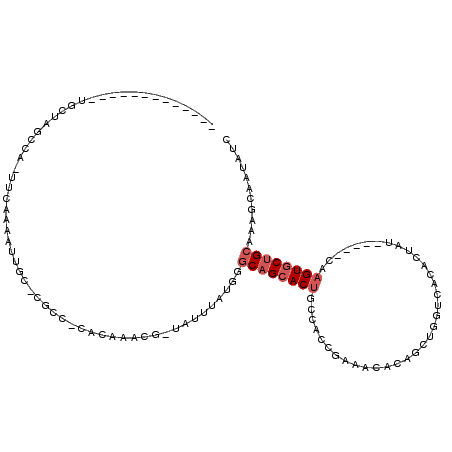
>X_DroMel_CAF1 10609628 98 - 22224390 -------------UGCUAGCCA-UUCAAAAUUGC-CGCC-CACAAACG-UUUUUAUGGGCAGCACUGUCAUCAAAACACAGCUGGUCACACCAU-----CAAGUGCUGCAAAGCAAUAUC -------------((((..(((-(..(((.(((.-....-..)))...-)))..))))((((((((((.......))...(.(((.....))).-----).))))))))..))))..... ( -22.60) >DroSec_CAF1 6770 99 - 1 -------------UGCUAGCCAAUUCAAAAUUGC-CGCC-CACAAACG-UUUUUAUGGGCAGCACUGCCAUCGAAACAUUGCUGGUCACACUUU-----CAAGUUCAGCAAAGCAAUAUC -------------((((..............(((-.(((-((.(((..-..))).))))).)))..............(((((((....(((..-----..))))))))))))))..... ( -23.00) >DroEre_CAF1 6742 97 - 1 -------------UGUU--CCA-UUCAAAACUAG-CGCC-CACAAGCUUUAAUUAUGGGCAGCACUGCCACCGAAACACGGCUGGUCGCACUAU-----CAAGUGCUGCAAAGCUAUAUC -------------....--...-........(((-((((-((.((.......)).))))).(((..(((((((.....))).)))).(((((..-----..))))))))...)))).... ( -29.20) >DroYak_CAF1 7331 97 - 1 -------------UGUCUGUCA-UUCAAAACUGG-CGCC-CAAAAGCU-CAUUAAUGGGCAGCACUGUCACCGAAACACGGCUGGUCACACUAU-----CGUGUGCUGCAAAGCUA-AUC -------------.........-........(((-((((-((......-......))))).(((...((((((.....))).))).(((((...-----.))))).)))...))))-... ( -23.20) >DroAna_CAF1 9050 109 - 1 CACUAAGUAGCAAGUCUAGCUA-UUUUGAAUUGCGCGCCUUACCAUCC-ACUUGAUGGGCAGCACUGGC--CUUAACAAAAUUGGCCACACUUUUUGGCCAAGUGCUGGACAG------- ..(.(((((((.......))))-))).)..............(((((.-....))))).((((((((((--(.....((((.((....)).)))).)))).))))))).....------- ( -31.40) >consensus _____________UGCUAGCCA_UUCAAAAUUGC_CGCC_CACAAACG_UAUUUAUGGGCAGCACUGCCACCGAAACACAGCUGGUCACACUAU_____CAAGUGCUGCAAAGCAAUAUC ..........................................................((((((((...................................))))))))........... ( -8.09 = -8.89 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:08 2006