
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,574,046 – 10,574,152 |
| Length | 106 |
| Max. P | 0.999171 |

| Location | 10,574,046 – 10,574,152 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.42 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.57 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10574046 106 + 22224390 CGGUGGACGGUGGGCGGUGGGCAGUGGGUGGCGGGGCGAUG-------------UACACAUGUAUGUAAAAAUGU-AAAGUGUACACUUAUUGGGUGGGGAAAAAACCUUGCCGACCGCU ((.....))...((((((...((((((((.((...))...(-------------(((((...(((((....))))-)..))))))))))))))((..(((......)))..)).)))))) ( -35.00) >DroSec_CAF1 14068 109 + 1 CGGUGG----------GUGGUCAGUGGGUGGCGGGGCGAUGGGAGUUACAUACAUACAAACAUAUGUAAAAAAGU-AAAGUGUACACUCAUUGGGUGGGGGAAAAACCUUGCCGACCGCU .....(----------(((((((((((((((((..((.......))..).((((((......)))))).......-.....)).)))))))).((..(((......)))..))))))))) ( -37.30) >DroSim_CAF1 13424 100 + 1 CGGUGG----------GUGGGCAGUGGGUGGCGGU---------GGUACAUACAUACAAACAUAUGUAAAAAAGU-AAAGUGUACACUCAUUGGGUGGGGGAGAAACCUUGCCGACCGCU .....(----------((((.((((((((((((.(---------..(((.((((((......)))))).....))-).).))).)))))))))((..(((......)))..))..))))) ( -34.90) >DroEre_CAF1 14046 82 + 1 CGGUGG----------GCGG----UGGGAGGU-----------------------AUGUAUGUACGUAAAAGUGG-AAGGUGUGCACUUAUUGGGUGGGGGAAAAACCUUGCCGACCACU .(((((----------.(((----((((..((-----------------------(..(.(.(((......))).-)..)..))).)))))))((..(((......)))..))..))))) ( -28.20) >DroYak_CAF1 15328 100 + 1 CGGUGG----------GUGG----UGGGAGGUGGUUAGGU----GGCAUGUAC-UAUGUACAUAUGUAAAAAUACAAAAGUGUACACUUAUUGGGCAGGGGAA-AACCUUGCCGACCGCU .....(----------((((----(..........(((((----(((((((((-...))))...((((....))))...)))).))))))...(((((((...-..))))))).)))))) ( -31.50) >consensus CGGUGG__________GUGG_CAGUGGGUGGCGGG__G_U____G__A__UAC_UACAAACAUAUGUAAAAAUGU_AAAGUGUACACUUAUUGGGUGGGGGAAAAACCUUGCCGACCGCU .(((((...............(((((((((......................................................)))))))))(((((((......)))))))..))))) (-18.98 = -19.86 + 0.88)



| Location | 10,574,046 – 10,574,152 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.42 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -6.96 |
| Energy contribution | -7.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10574046 106 - 22224390 AGCGGUCGGCAAGGUUUUUUCCCCACCCAAUAAGUGUACACUUU-ACAUUUUUACAUACAUGUGUA-------------CAUCGCCCCGCCACCCACUGCCCACCGCCCACCGUCCACCG .(((((.((((.((........)).........((((((((...-................)))))-------------)))...............)))).)))))............. ( -24.61) >DroSec_CAF1 14068 109 - 1 AGCGGUCGGCAAGGUUUUUCCCCCACCCAAUGAGUGUACACUUU-ACUUUUUUACAUAUGUUUGUAUGUAUGUAACUCCCAUCGCCCCGCCACCCACUGACCAC----------CCACCG .((((..(((..((........)).......(((((((.....)-)).....(((((((((....))))))))))))).....)))))))..............----------...... ( -20.90) >DroSim_CAF1 13424 100 - 1 AGCGGUCGGCAAGGUUUCUCCCCCACCCAAUGAGUGUACACUUU-ACUUUUUUACAUAUGUUUGUAUGUAUGUACC---------ACCGCCACCCACUGCCCAC----------CCACCG .(.(((.((((.(((................(((((.......)-))))...(((((((((....)))))))))..---------......)))...)))).))----------)).... ( -19.50) >DroEre_CAF1 14046 82 - 1 AGUGGUCGGCAAGGUUUUUCCCCCACCCAAUAAGUGCACACCUU-CCACUUUUACGUACAUACAU-----------------------ACCUCCCA----CCGC----------CCACCG .((((.(((..((((................(((((........-.)))))..............-----------------------))))....----))).----------)))).. ( -14.71) >DroYak_CAF1 15328 100 - 1 AGCGGUCGGCAAGGUU-UUCCCCUGCCCAAUAAGUGUACACUUUUGUAUUUUUACAUAUGUACAUA-GUACAUGCC----ACCUAACCACCUCCCA----CCAC----------CCACCG ...(((.((((.(((.-.......)))......(((((((....((((....))))..))))))).-.....))))----))).............----....----------...... ( -21.30) >consensus AGCGGUCGGCAAGGUUUUUCCCCCACCCAAUAAGUGUACACUUU_ACAUUUUUACAUAUGUAUGUA_GUA__U__C____A_C__ACCGCCACCCACUG_CCAC__________CCACCG ...((..(((..(((.........)))....((((....)))).............................................)))..))......................... ( -6.96 = -7.16 + 0.20)

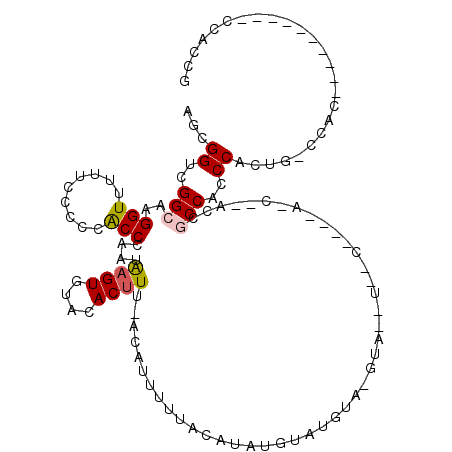
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:46 2006