
| Sequence ID | X_DroMel_CAF1 |
|---|---|
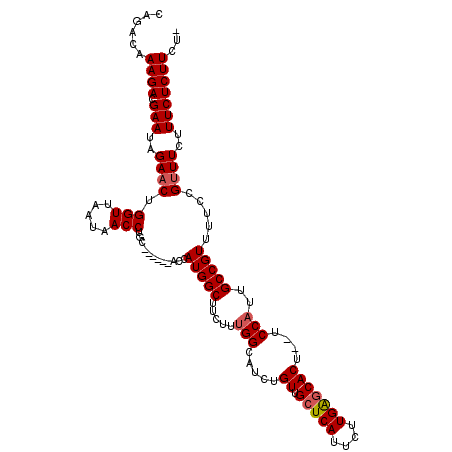
| Location | 10,566,431 – 10,566,542 |
| Length | 111 |
| Max. P | 0.888097 |

| Location | 10,566,431 – 10,566,542 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -20.49 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10566431 111 + 22224390 CAGACAAAGACGAAUAGAACUGGUUAAUAACCUGCGCA--CGCCAUGGCUUCCUUGGCAUCUGUUGCUCAUUCUUGGCCACU--UCCAUUGCCGUUUUCCGUUUCUUUCUCUUCU- .((((.(((((((((.(((.(((((((......(((((--.((((.((...)).))))...)).)))......))))))).)--)).)))..))))))..))))...........- ( -23.20) >DroSec_CAF1 6594 113 + 1 CAGACAAAGACGAAUAGAACUGGUUAAUAACCUGCGCACACACCAUGGCUUCUUCGGCAUCUGUUGCUCAUUCUUGAGCACU--UCCAUUGCCGUUAUCCGUUUCUUUCUCUUCU- ......((((.(((.((((.(((.((((.......((.((.....))))......((((.....((((((....))))))..--.....)))))))).))).)))))))))))..- ( -25.90) >DroEre_CAF1 6542 108 + 1 CAGACAAAGACGAAUAGAACUGGUUAAUAACCUGC------ACCAUGGCUUCUUUGGCAUCUGUUGCUCAUUCUUGAGCACU--UCCAUUGCCGUUUUCCGUUUCUUUCUCUUCUG ((((...(((.(((.((((.(((..(((..((...------.....)).......((((.....((((((....))))))..--.....)))))))..))).)))))))))))))) ( -26.20) >DroYak_CAF1 7202 110 + 1 CAGACAAAGACGAAUAGAACUGGUUAAUAACCUGC------AGCAUGGCUUCUUUGGCAUCUGUUGCUCAUUCUUGAGCACUUUUCCAUUGCCGUUUUCCGCUUCUUUCUCUUCUU .(((.(((((.(...(((((.(((.........((------((.(((.(......).)))))))((((((....))))))..........))))))))...).))))).))).... ( -25.70) >consensus CAGACAAAGACGAAUAGAACUGGUUAAUAACCUGC______ACCAUGGCUUCUUUGGCAUCUGUUGCUCAUUCUUGAGCACU__UCCAUUGCCGUUUUCCGUUUCUUUCUCUUCU_ ......((((.(((..((((.(((.....)))............(((((.....(((.....((.(((((....)))))))....)))..))))).....))))..)))))))... (-20.49 = -21.05 + 0.56)


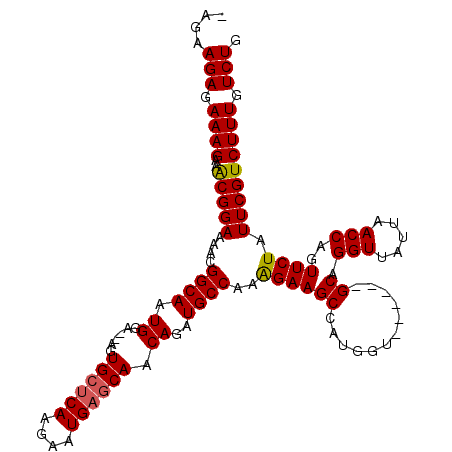
| Location | 10,566,431 – 10,566,542 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -24.73 |
| Energy contribution | -24.85 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10566431 111 - 22224390 -AGAAGAGAAAGAAACGGAAAACGGCAAUGGA--AGUGGCCAAGAAUGAGCAACAGAUGCCAAGGAAGCCAUGGCG--UGCGCAGGUUAUUAACCAGUUCUAUUCGUCUUUGUCUG -...(((.(((((..((.....))(.((((((--(.(((.....(((((.(..(..((((((.((...)).)))))--)..)..).)))))..))).)))))))).))))).))). ( -30.40) >DroSec_CAF1 6594 113 - 1 -AGAAGAGAAAGAAACGGAUAACGGCAAUGGA--AGUGCUCAAGAAUGAGCAACAGAUGCCGAAGAAGCCAUGGUGUGUGCGCAGGUUAUUAACCAGUUCUAUUCGUCUUUGUCUG -...(((.(((((..((((((.(((((.((..--..((((((....)))))).))..)))))..((((((((.....))).)).(((.....)))..)))))))))))))).))). ( -35.30) >DroEre_CAF1 6542 108 - 1 CAGAAGAGAAAGAAACGGAAAACGGCAAUGGA--AGUGCUCAAGAAUGAGCAACAGAUGCCAAAGAAGCCAUGGU------GCAGGUUAUUAACCAGUUCUAUUCGUCUUUGUCUG ....(((.(((((..((((....((((.((..--..((((((....)))))).))..))))..((((....((((------...........)))).)))).))))))))).))). ( -29.70) >DroYak_CAF1 7202 110 - 1 AAGAAGAGAAAGAAGCGGAAAACGGCAAUGGAAAAGUGCUCAAGAAUGAGCAACAGAUGCCAAAGAAGCCAUGCU------GCAGGUUAUUAACCAGUUCUAUUCGUCUUUGUCUG ....(((.(((((.((((.....(((..(((.....((((((....)))))).......))).....)))...))------)).(((.....)))...........))))).))). ( -28.60) >consensus _AGAAGAGAAAGAAACGGAAAACGGCAAUGGA__AGUGCUCAAGAAUGAGCAACAGAUGCCAAAGAAGCCAUGGU______GCAGGUUAUUAACCAGUUCUAUUCGUCUUUGUCUG ....(((.((((..(((((....((((.((......((((((....)))))).))..))))..((((((............)).(((.....)))..)))).))))))))).))). (-24.73 = -24.85 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:43 2006