
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,558,874 – 10,559,001 |
| Length | 127 |
| Max. P | 0.935023 |

| Location | 10,558,874 – 10,558,967 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.29 |
| Mean single sequence MFE | -11.70 |
| Consensus MFE | -9.49 |
| Energy contribution | -9.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10558874 93 - 22224390 AAAUUUUGGUAAACUAAAAUGGAAUUUAGAUUUAAUUUUAGAUACACCAAAUCAAAUCUAAUACAACACACACGAUCAC----ACACACACACACCU ....((((((...(((((((.((((....)))).)))))))....))))))............................----.............. ( -10.00) >DroSec_CAF1 85251 97 - 1 AAAUUUUGGUAAACUAAAAUGGAAUUUAGAUCUAGUUUUAGACACACCAAAUGAAAUCUAAUACAACACACACGAUCACGAUCACACACACACACCU ....((((((...((((((((((.......)))).))))))....))))))......................(((....))).............. ( -10.70) >DroSim_CAF1 89945 97 - 1 AAAUUUUGGUAAACUAAAAUGGAAUUUAGAUUUAGUUUUAGACACACCAAAUCAAAUCUAAUACAACACACACGGUCACGAUCACACACACACACCU ....((((((...(((((((.((((....)))).)))))))....)))))).....................((....))................. ( -11.10) >DroEre_CAF1 87041 88 - 1 GAAUUUUGGUAGACUAAAAUGGAAU-UAUAUUUCGUUUUAGACGCACCAAAUCAAAUCUAAUAACACACAC--------GAUCACACACACACACCU ....((((((.(.((((((((((((-...)))))))))))).)..))))))....................--------.................. ( -15.00) >consensus AAAUUUUGGUAAACUAAAAUGGAAUUUAGAUUUAGUUUUAGACACACCAAAUCAAAUCUAAUACAACACACACGAUCACGAUCACACACACACACCU ....((((((...(((((((.((((....)))).)))))))....)))))).............................................. ( -9.49 = -9.55 + 0.06)



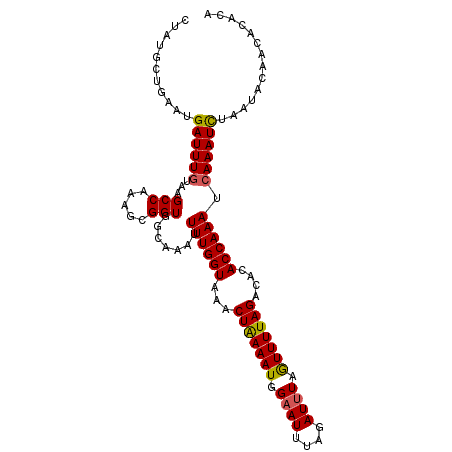
| Location | 10,558,895 – 10,559,001 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -18.10 |
| Energy contribution | -19.14 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10558895 106 + 22224390 UGUGUGUUGUAUUAGAUUUGAUUUGGUGUAUCUAAAAUUAAAUCUAAAUUCCAUUUUAGUUUACCAAAAUUUGCCACCGCUUUGGCUUACAAAUCAUUCAGCAUAG ..(((((((.....((((((.(((((((...(((((((..(((....)))..)))))))..)))))))....((((......))))...))))))...))))))). ( -27.70) >DroSec_CAF1 85276 106 + 1 UGUGUGUUGUAUUAGAUUUCAUUUGGUGUGUCUAAAACUAGAUCUAAAUUCCAUUUUAGUUUACCAAAAUUUGCCACCGCUUUGGCUUACAAAUCAUUCAGCAUAG ..(((((((.....(((((..(((((((.(((((....)))))(((((......)))))..)))))))....((((......))))....)))))...))))))). ( -23.20) >DroSim_CAF1 89970 106 + 1 UGUGUGUUGUAUUAGAUUUGAUUUGGUGUGUCUAAAACUAAAUCUAAAUUCCAUUUUAGUUUACCAAAAUUUGCCACCGCUUUGGCUUACAAAUCAUUCAGCAUAG ..(((((((.....((((((.(((((((...((((((...(((....)))...))))))..)))))))....((((......))))...))))))...))))))). ( -25.50) >DroEre_CAF1 87059 104 + 1 -GUGUGUGUUAUUAGAUUUGAUUUGGUGCGUCUAAAACGAAAUAUA-AUUCCAUUUUAGUCUACCAAAAUUCGUCACCGCUUUGGCUUACAAAUCAUUCAGCAUAG -...((((((....((((((.(((((((.(.((((((.(((.....-.)))..)))))).))))))))....((((......))))...))))))....)))))). ( -23.20) >DroYak_CAF1 89332 105 + 1 UGUGUGUGUUAUUAAAUUUGAUUUGGUGUGUCUAAAAUUAAAUAUA-AUUCCAUUUCAGUUUACCAAAAUUUUUCACCGCUUUGGCUUACAAAUCAAUUAGUAUAG .........((((((..(((((((((.(((...((((((......(-(((.......))))......)))))).))))((....))...))))))))))))))... ( -12.60) >consensus UGUGUGUUGUAUUAGAUUUGAUUUGGUGUGUCUAAAACUAAAUCUAAAUUCCAUUUUAGUUUACCAAAAUUUGCCACCGCUUUGGCUUACAAAUCAUUCAGCAUAG ..(((((((.....((((((.(((((((...((((((...(((....)))...))))))..)))))))....((((......))))...))))))...))))))). (-18.10 = -19.14 + 1.04)

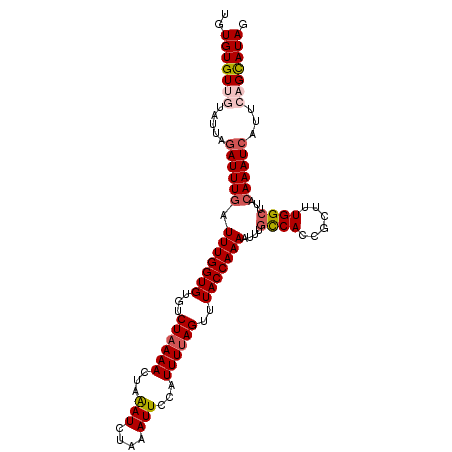

| Location | 10,558,895 – 10,559,001 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10558895 106 - 22224390 CUAUGCUGAAUGAUUUGUAAGCCAAAGCGGUGGCAAAUUUUGGUAAACUAAAAUGGAAUUUAGAUUUAAUUUUAGAUACACCAAAUCAAAUCUAAUACAACACACA ...........((((((...((((......))))....((((((...(((((((.((((....)))).)))))))....)))))).)))))).............. ( -20.30) >DroSec_CAF1 85276 106 - 1 CUAUGCUGAAUGAUUUGUAAGCCAAAGCGGUGGCAAAUUUUGGUAAACUAAAAUGGAAUUUAGAUCUAGUUUUAGACACACCAAAUGAAAUCUAAUACAACACACA ..............(((((.((((......))))....((((((...((((((((((.......)))).))))))....))))))..........)))))...... ( -17.90) >DroSim_CAF1 89970 106 - 1 CUAUGCUGAAUGAUUUGUAAGCCAAAGCGGUGGCAAAUUUUGGUAAACUAAAAUGGAAUUUAGAUUUAGUUUUAGACACACCAAAUCAAAUCUAAUACAACACACA ...........((((((...((((......))))....((((((...(((((((.((((....)))).)))))))....)))))).)))))).............. ( -20.60) >DroEre_CAF1 87059 104 - 1 CUAUGCUGAAUGAUUUGUAAGCCAAAGCGGUGACGAAUUUUGGUAGACUAAAAUGGAAU-UAUAUUUCGUUUUAGACGCACCAAAUCAAAUCUAAUAACACACAC- ..........(((((((...((((((.((....))...))))))...((((((((((((-...))))))))))))......))))))).................- ( -25.60) >DroYak_CAF1 89332 105 - 1 CUAUACUAAUUGAUUUGUAAGCCAAAGCGGUGAAAAAUUUUGGUAAACUGAAAUGGAAU-UAUAUUUAAUUUUAGACACACCAAAUCAAAUUUAAUAACACACACA .........(((((((....((....))((((.....(((..(....)..)))((((((-((....))))))))....)))))))))))................. ( -17.20) >consensus CUAUGCUGAAUGAUUUGUAAGCCAAAGCGGUGGCAAAUUUUGGUAAACUAAAAUGGAAUUUAGAUUUAGUUUUAGACACACCAAAUCAAAUCUAAUACAACACACA ...........((((((...(((.....))).......((((((...(((((((.((((....)))).)))))))....)))))).)))))).............. (-15.46 = -15.30 + -0.16)

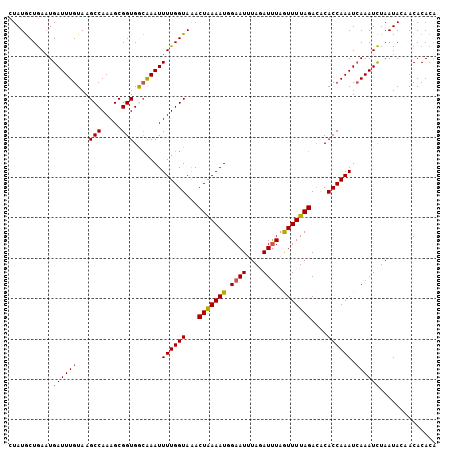
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:38 2006