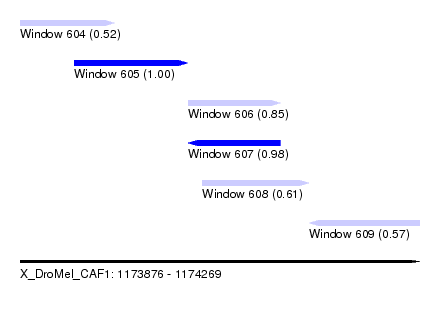
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,173,876 – 1,174,269 |
| Length | 393 |
| Max. P | 0.999668 |

| Location | 1,173,876 – 1,173,969 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 98.57 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -37.53 |
| Energy contribution | -37.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 1.00 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1173876 93 + 22224390 ACCCCUCGAAAUGGGGAACGCAGCCGUGCAACCGAGCGGCUGGUUGGCUGCUGAAAGGUCAUUGGGCUUGUUCAGACGCCCGUUCAGCUCAGA ..(((.......)))((((((((((((........)))))))...(((..((((((((((....))))).)))))..))))))))........ ( -36.60) >DroSec_CAF1 9893 93 + 1 ACCCCUCGAAAUGGGGAACGCAGCCGUGCAACGGGGCGGCUGGUUGGCUGCUGAAAGGUCAUUGGGCUUGUUCAGACGCCCGUUCAGCUCAGA ..(((.......)))((((((((((((.(....).)))))))...(((..((((((((((....))))).)))))..))))))))........ ( -38.20) >DroSim_CAF1 24981 93 + 1 ACCCCUCGAAAUGGGGAACGCAGCCGUGCAACGGGGCGGCUGGUUGGCUGCUGAAAGGUCAUUGGGCUUGUUCAGACGCCCGUUCAGCUCAGA ..(((.......)))((((((((((((.(....).)))))))...(((..((((((((((....))))).)))))..))))))))........ ( -38.20) >consensus ACCCCUCGAAAUGGGGAACGCAGCCGUGCAACGGGGCGGCUGGUUGGCUGCUGAAAGGUCAUUGGGCUUGUUCAGACGCCCGUUCAGCUCAGA ..(((.......)))((((((((((((.(....).)))))))...(((..((((((((((....))))).)))))..))))))))........ (-37.53 = -37.53 + 0.00)

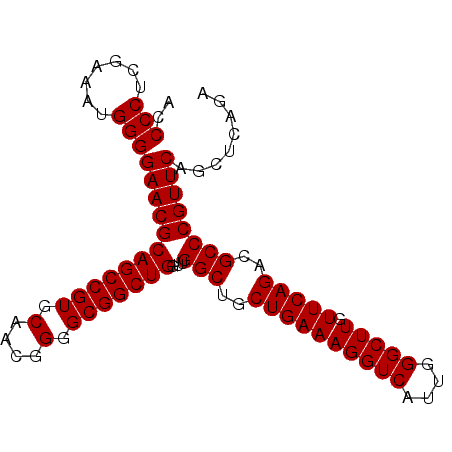
| Location | 1,173,929 – 1,174,041 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.55 |
| Mean single sequence MFE | -45.14 |
| Consensus MFE | -39.18 |
| Energy contribution | -38.86 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1173929 112 + 22224390 AAAGGUCAUUGGGCUUGUUCAGACGCCCGUUCAGCUCAGACGGAGUUCGAAGCUGCAUCCUGCUUGAGCCCUUGCUGC--------UCCGUGUCAAUGACACGCCCUUUGGCCAAUACCC ...(((((..((((.((((..(((((..(..((((......((.((((.((((........))))))))))..)))).--------.).)))))...)))).))))..)))))....... ( -41.80) >DroSec_CAF1 9946 112 + 1 AAAGGUCAUUGGGCUUGUUCAGACGCCCGUUCAGCUCAGACGGAGUUCGGAGCUGCAUCCUGCUUGAGCCCUUGCUGC--------UCCGUGUCAAUGACACGCCCUUUGGCCAAACCCC ...(((((..((((.((((..((((((((((.......))))).....(((((.(((....((....))...))).))--------))))))))...)))).))))..)))))....... ( -47.70) >DroSim_CAF1 25034 112 + 1 AAAGGUCAUUGGGCUUGUUCAGACGCCCGUUCAGCUCAGACGGAGUUCGGAGCUGCAUCCUGCUUGAGCCCUUGCUGC--------UCCGUGUCAAUGACACGCCCUUUGGCCAAACCCC ...(((((..((((.((((..((((((((((.......))))).....(((((.(((....((....))...))).))--------))))))))...)))).))))..)))))....... ( -47.70) >DroEre_CAF1 9115 104 + 1 AAAGGUCAUUGGGCUUGUUCAGACGCCCGUUC-----AGACGGAGUUUGGAGCUGCAUCCUGCUUGAGCAUUUGCUGC--------UCAGUGUCAAUGACACGCCCUUUGGCC---CCUC ...(((((..((((.....(((((..((((..-----..)))).)))))((((.(((...(((....)))..))).))--------)).(((((...)))))))))..)))))---.... ( -45.00) >DroYak_CAF1 11924 111 + 1 AAAGGUCAUUGGGCUUGUUCAGACGCCCGUUC-----AGACGGAGUUCGGAGCUGCAUCCUGCUUGAGCCUCUACUGCACUGAGGCGCAGUGUCAAUGACACGCCCUUUGGCC----CUC ...(((((..((((.((((..(((((((((..-----..))))(((..(((......))).)))((.(((((.........))))).)))))))...)))).))))..)))))----... ( -43.50) >consensus AAAGGUCAUUGGGCUUGUUCAGACGCCCGUUCAGCUCAGACGGAGUUCGGAGCUGCAUCCUGCUUGAGCCCUUGCUGC________UCCGUGUCAAUGACACGCCCUUUGGCCAA_CCCC ...(((((..((((.((((..((((((((((.......))))).((((.((((........))))))))....................)))))...)))).))))..)))))....... (-39.18 = -38.86 + -0.32)

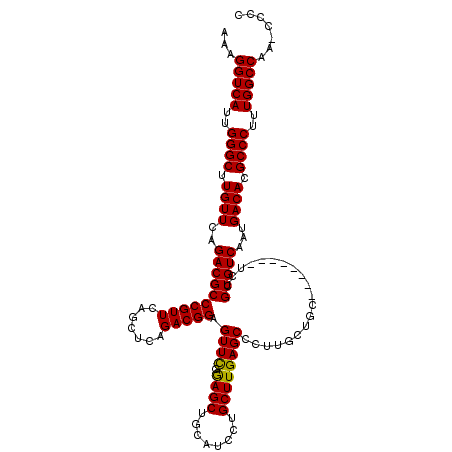
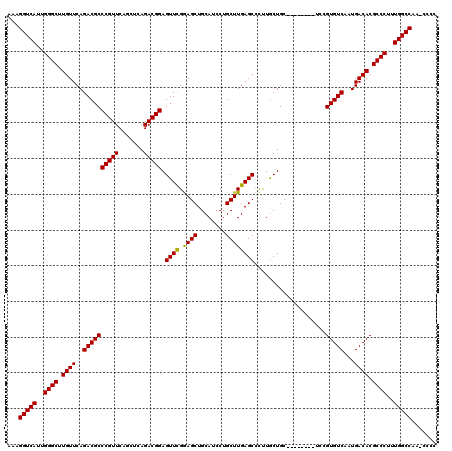
| Location | 1,174,041 – 1,174,132 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -31.76 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1174041 91 + 22224390 CGUCGCCCCACCGG-CGCUCAUUAGCGCAGCUUUAGGGCCAAUGCCAAAGGUGGGCAAAGAAAGCCCGAGGUGGGGACAAAGCCUCUGCAGC ....((((((((.(-((((....))))).(((((..(((....))).)))))((((.......))))..))))))).)...((....))... ( -37.30) >DroSec_CAF1 10058 92 + 1 CGCCGCCCCACCUGCCGCUCAUUAGCGCAGCUUUGGGGCCAAUGCCAAAGGCGGGCAAGGAAAGCCCGAGGUGGGGACAAAGCCUCUGCAGC .((.((((((.((((.(((....)))))))...))))))...(((...((((((((.......))))....((....))..))))..))))) ( -38.50) >DroSim_CAF1 25146 92 + 1 CGCCGCCCCACCUGCCGCUCAUUAGCGCAGCUUUAGGGCCAAUGCCAAAGGCGGGCAAGGAAAGCCCGAGGUGGGGACAAAGCCUCUGCAGC .((.(((((((((((.(((....))))).(((((..(((....))).)))))((((.......)))).)))))))).)...((....)).)) ( -37.00) >DroEre_CAF1 9219 92 + 1 CGCUGCCCCACCUGCCGCUCAUUAGCGCAGCUUUAAGGCCAGCGCCAAAGGCAGGCGAACAAAGCCCCAGAUGGGGACAAAACCUCUGCAGC .(((((..(.((((((........((((.(((....)))..))))....)))))).)......(((((....)))).).........))))) ( -35.10) >consensus CGCCGCCCCACCUGCCGCUCAUUAGCGCAGCUUUAGGGCCAAUGCCAAAGGCGGGCAAAGAAAGCCCGAGGUGGGGACAAAGCCUCUGCAGC ....((((((((((.((((....))))).(((((..(((....))).)))))((((.......)))).)))))))).).............. (-31.76 = -32.33 + 0.56)

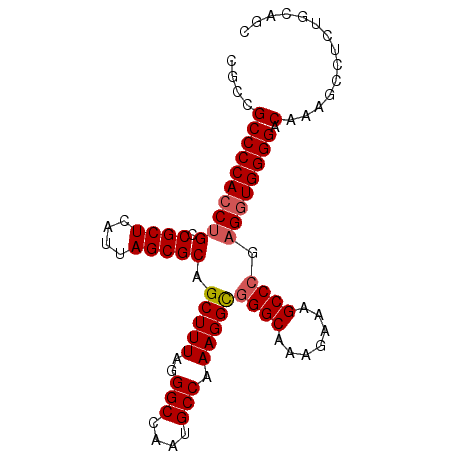
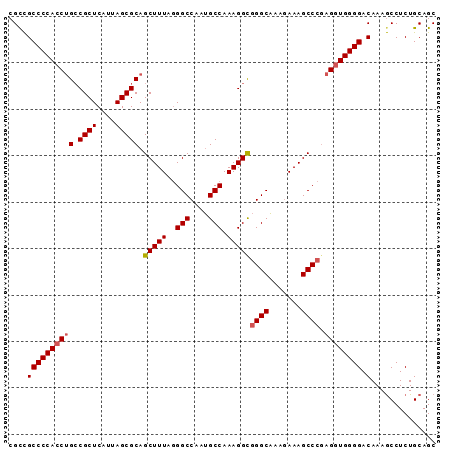
| Location | 1,174,041 – 1,174,132 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -39.12 |
| Energy contribution | -38.50 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1174041 91 - 22224390 GCUGCAGAGGCUUUGUCCCCACCUCGGGCUUUCUUUGCCCACCUUUGGCAUUGGCCCUAAAGCUGCGCUAAUGAGCG-CCGGUGGGGCGACG (((.....))).(((.(((((((..(((((.....((((.......))))..))))).......(((((....))))-).)))))))))).. ( -39.50) >DroSec_CAF1 10058 92 - 1 GCUGCAGAGGCUUUGUCCCCACCUCGGGCUUUCCUUGCCCGCCUUUGGCAUUGGCCCCAAAGCUGCGCUAAUGAGCGGCAGGUGGGGCGGCG (((((...(((...)))(((((((.(((((.....((((.......))))..)))))....(((((........))))))))))))))))). ( -43.60) >DroSim_CAF1 25146 92 - 1 GCUGCAGAGGCUUUGUCCCCACCUCGGGCUUUCCUUGCCCGCCUUUGGCAUUGGCCCUAAAGCUGCGCUAAUGAGCGGCAGGUGGGGCGGCG (((((...(((...)))(((((((.(((((.....((((.......))))..)))))....(((((........))))))))))))))))). ( -44.00) >DroEre_CAF1 9219 92 - 1 GCUGCAGAGGUUUUGUCCCCAUCUGGGGCUUUGUUCGCCUGCCUUUGGCGCUGGCCUUAAAGCUGCGCUAAUGAGCGGCAGGUGGGGCAGCG (((((.........(((((.....)))))....((((((((((.(((((((.(((......)))))))))).....))))))))))))))). ( -45.30) >consensus GCUGCAGAGGCUUUGUCCCCACCUCGGGCUUUCCUUGCCCGCCUUUGGCAUUGGCCCUAAAGCUGCGCUAAUGAGCGGCAGGUGGGGCGGCG (((((...(((...)))((((((..(((((.....((((.......))))..)))))....(((((........))))).))))))))))). (-39.12 = -38.50 + -0.62)

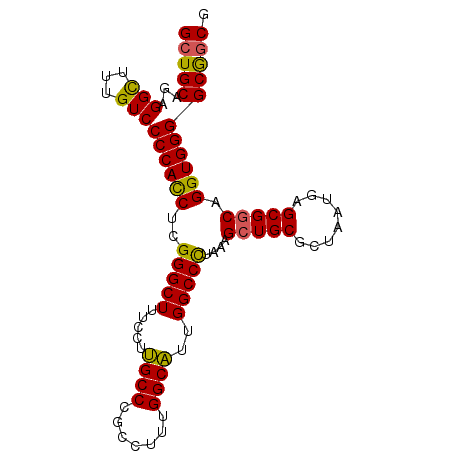
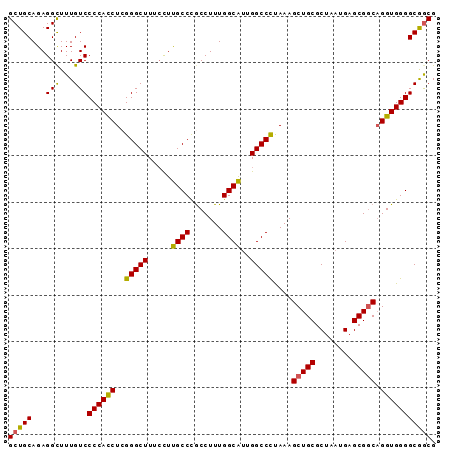
| Location | 1,174,055 – 1,174,160 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.97 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1174055 105 + 22224390 ---CGCUCAUUAGCGCAGCUUUAGGGCCAAUGCCAAAGGUGGGCAAAGAAAGCCCGAGGUGGGGACAAAGCCUCUGCAGCGAAAA------------ACAUGCAAGGCUCGGACCAAAAA ---.(((..((.((.((.(((...(((....))).))).)).)).))...)))((((..((....))..((((.((((.......------------...))))))))))))........ ( -31.80) >DroSec_CAF1 10072 106 + 1 --CCGCUCAUUAGCGCAGCUUUGGGGCCAAUGCCAAAGGCGGGCAAGGAAAGCCCGAGGUGGGGACAAAGCCUCUGCAGCGAGAA------------ACAUGCAAGGCGCGGACCAAAAA --((((.......(((.(((((..(((....))).)))))((((.......))))...)))........((((.((((.......------------...))))))))))))........ ( -36.80) >DroYak_CAF1 12075 106 + 1 AGCCGCUCAUUAGUGCAGCUUUGGGGCCAGCGCCAAAGGCAGCCAA----AGC----------AACACAACCUCUGCAGCGAAUGGCACGUAGAAGAAAAUGCGCAACGCGGACCAAAAA ..((((......((((((((((((.(((.........)))..))))----)))----------.......(.(((((.((.....))..))))).)....)))))...))))........ ( -33.20) >consensus __CCGCUCAUUAGCGCAGCUUUGGGGCCAAUGCCAAAGGCGGGCAA_GAAAGCCCGAGGUGGGGACAAAGCCUCUGCAGCGAAAA____________ACAUGCAAGGCGCGGACCAAAAA ...((((((...((...(((((..(((....))).)))))((((.......))))..............))...)).))))....................................... (-18.50 = -18.83 + 0.34)

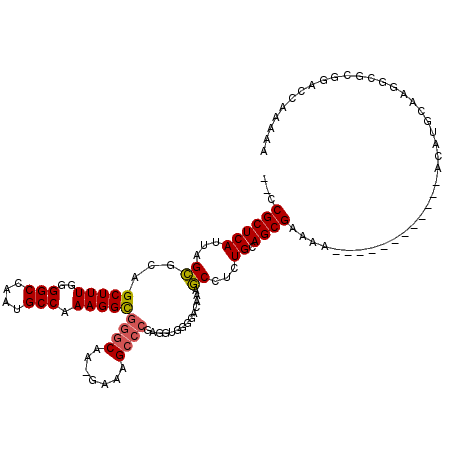
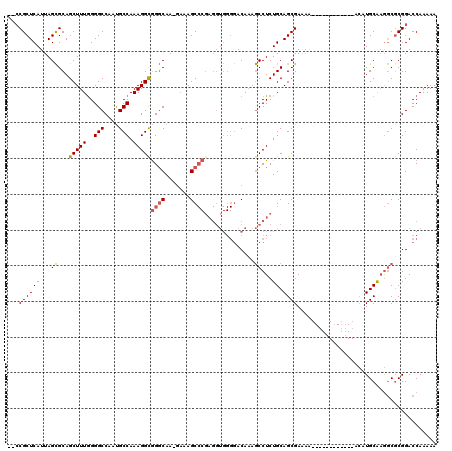
| Location | 1,174,160 – 1,174,269 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -18.83 |
| Energy contribution | -20.14 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1174160 109 - 22224390 UCGCAAAUAUUUGCUCAUUUCGCAAUGUGUGGAUAGCUGCGU---GAAGGGGCUAA-AAGCCUCUUAUCGCUGCCCAAUGUUUGUGCGGCCAGC-------UACAUAUUUGAGAUUCGCA ...........(((..((((((.(((((((((...((((((.---.((.((((...-.(((........)))))))....))..))))))...)-------))))))))))))))..))) ( -37.20) >DroSim_CAF1 25266 116 - 1 UCGCAAAUAUUUGCUCAUUUCGCAAUGUGGGGAUAGCUACGC---GAAGGGGCUGA-AAGCCUCUUAUCGCUGCCCAAUGUUUGUGCGGCCAGCAGGCCUCCACAUAUUUGAGAUUCGCA ((.((((((.((((.......))))(((((((...(((..((---.(((((((...-..)))))))...(((((.((.....)).)))))..)).)))))))))))))))).))...... ( -38.30) >DroEre_CAF1 9350 98 - 1 UCGCAAAUAUUUCCUCAUUU--------CUGUAUUGCUGG-----AAAGGGGCUGA-AAGCCUCUUAUCGGUGGCAAAUGUUUGAGCGGGCAGAGA--------UUAUUUGAGAUUCGCA ..((....(((((...((((--------((((.(..((((-----.(((((((...-..))))))).))))..)....(((....))).)))))))--------).....)))))..)). ( -28.30) >DroYak_CAF1 12181 116 - 1 UCGCAAAUAUUUGCCCAUUUCGUCAUGUCGGGAUUGCUGGGGGAUAAAGGGGCUAAGAAGCCUCUUAUCGCUGGCAAAUGUUUGAGCGGCCAGCGAGAAG----CUAUUUGAGCUUCGCA ..((......((.(((((..((......))..)....)))).))..((((((((....)))))))).((((((((....((....)).))))))))((((----((.....)))))))). ( -44.30) >consensus UCGCAAAUAUUUGCUCAUUUCGCAAUGUCGGGAUAGCUGCG____AAAGGGGCUAA_AAGCCUCUUAUCGCUGCCAAAUGUUUGAGCGGCCAGCGA________AUAUUUGAGAUUCGCA ...((((((((((((.............((((....))))......((((((((....))))))))......)))))))))))).((((..........................)))). (-18.83 = -20.14 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:36 2006