
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,538,291 – 10,538,489 |
| Length | 198 |
| Max. P | 0.999446 |

| Location | 10,538,291 – 10,538,411 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.84 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10538291 120 + 22224390 CCAGAAAAAAACCCAAUACUCCCACUCCACCGACCCAUUUUGAACAGAGAGAAAAAAAUGUAGUUAUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCA .(((((((.....((((.......(((...(((......)))....))).....(((((((((.((((((((..(....)..)))))))).)))))))))))))......)))))))... ( -22.70) >DroSec_CAF1 64730 110 + 1 CCAGAAAAAAACCCAAUACCCCCACUCCACCA---------UAACAGAGAGAAA-AAAUGUAGUUAUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCA .(((((((.....((((.......((((....---------.....).)))..(-((((((((.((((((((..(....)..)))))))).)))))))))))))......)))))))... ( -22.50) >DroSim_CAF1 68548 110 + 1 GCAGAAAAAAACCCAAUACCCCCACUCCACCA---------UAACAGAGAGAAA-AAAUGUAGUUAUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCA ((((((((.....((((.......((((....---------.....).)))..(-((((((((.((((((((..(....)..)))))))).)))))))))))))......)))))))).. ( -26.00) >DroEre_CAF1 66150 94 + 1 CCAAAAAAAAAAAAAAGAG-------------------------CAGGGAGAAA-AAAUGUAGCUAUGGCUUAAAGCAAUACAAGCCAUAUUUAACUUUUAUUGACCAUGUUUUUUGCCA .............((((((-------------------------((((..((.(-(((..(((.((((((((..(....)..)))))))).)))..)))).))..)).)))))))).... ( -20.60) >DroYak_CAF1 67140 105 + 1 GCAGAAAAAAACCAAAUACCCCCC------AGA---------CACAGAGAAAAAAAUAUGUAGUUUUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCA ((((((((......((((......------...---------(.....)........((((((...((((((..(....)..))))))...))))))..)))).......)))))))).. ( -15.92) >consensus CCAGAAAAAAACCCAAUACCCCCACUCCACCA__________AACAGAGAGAAA_AAAUGUAGUUAUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCA ............................................(((((((....((((((((.((((((((..........)))))))).))))))))...........)))))))... (-14.36 = -14.84 + 0.48)

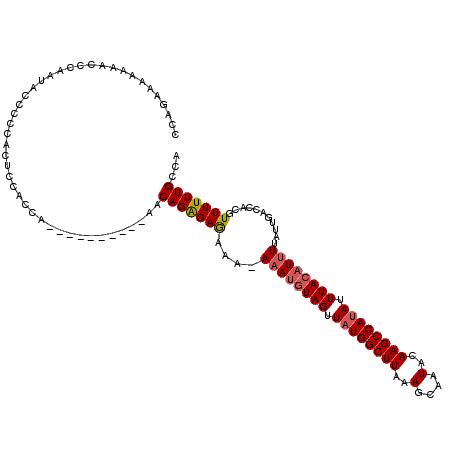

| Location | 10,538,331 – 10,538,449 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10538331 118 + 22224390 UGAACAGAGAGAAAAAAAUGUAGUUAUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAAUCUGCCUUGUAU--GUCAAUACACAAUGCGGUUGGAAAU ....(((((((...(((((((((.((((((((..(....)..)))))))).)))))))))..........)))))))((((.((((..(((((--....)))))....)))))))).... ( -29.72) >DroSec_CAF1 64762 116 + 1 -UAACAGAGAGAAA-AAAUGUAGUUAUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAGUCUGCCUUGUAU--GUCAAUACACAAUGCGGUUGGAAAU -...(((((((..(-((((((((.((((((((..(....)..)))))))).)))))))))..........)))))))((((.((((..(((((--....)))))....)))))))).... ( -29.80) >DroSim_CAF1 68580 116 + 1 -UAACAGAGAGAAA-AAAUGUAGUUAUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAGUCUGCCUUGUAU--GUCAAUACACAAUGCGGUUGGAAAU -...(((((((..(-((((((((.((((((((..(....)..)))))))).)))))))))..........)))))))((((.((((..(((((--....)))))....)))))))).... ( -29.80) >DroEre_CAF1 66169 113 + 1 ----CAGGGAGAAA-AAAUGUAGCUAUGGCUUAAAGCAAUACAAGCCAUAUUUAACUUUUAUUGACCAUGUUUUUUGCCAGUCUGCCUUGUAU--GUCAAUACACAAUGCGGCUGGAAAU ----..((..((.(-(((..(((.((((((((..(....)..)))))))).)))..)))).))..))..........((((.((((..(((((--....)))))....)))))))).... ( -28.90) >DroYak_CAF1 67167 118 + 1 --CACAGAGAAAAAAAUAUGUAGUUUUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAGUCUGCCUUGUGUUGGCAAAUACACAAUGCGGUUGGAAAU --..(((((((...(((((((((...((((((..(....)..))))))...)))))...)))).......)))))))((((.((((.((((((........)))))).)))))))).... ( -29.80) >consensus __AACAGAGAGAAA_AAAUGUAGUUAUGGCUUAAAGCAAUACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAGUCUGCCUUGUAU__GUCAAUACACAAUGCGGUUGGAAAU ....(((((((....((((((((.((((((((..........)))))))).))))))))...........)))))))((((.((((.((((............)))).)))))))).... (-25.94 = -26.10 + 0.16)


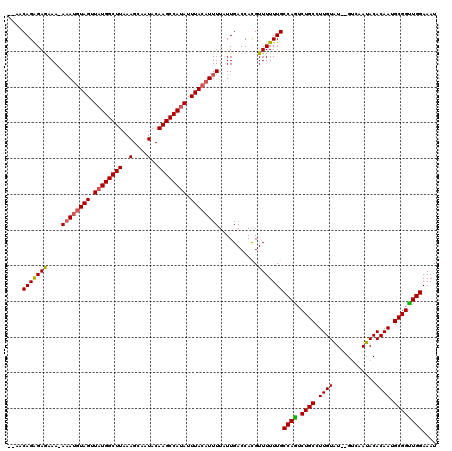
| Location | 10,538,331 – 10,538,449 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -24.26 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10538331 118 - 22224390 AUUUCCAACCGCAUUGUGUAUUGAC--AUACAAGGCAGAUUGGCAAAAAACGUGGUCAAUAAAAUGUAAAUAUGGCUUGUAUUGCUUUAAGCCAUAACUACAUUUUUUUCUCUCUGUUCA .((((((((.((.((((((......--)))))).)).).)))).))).((((.((..((.((((((((..(((((((((........)))))))))..)))))))).))..)).)))).. ( -30.70) >DroSec_CAF1 64762 116 - 1 AUUUCCAACCGCAUUGUGUAUUGAC--AUACAAGGCAGACUGGCAAAAAACGUGGUCAAUAAAAUGUAAAUAUGGCUUGUAUUGCUUUAAGCCAUAACUACAUUU-UUUCUCUCUGUUA- .((((((..(((.((((((......--)))))).)).)..))).))).((((.((.....((((((((..(((((((((........)))))))))..)))))))-)....)).)))).- ( -28.80) >DroSim_CAF1 68580 116 - 1 AUUUCCAACCGCAUUGUGUAUUGAC--AUACAAGGCAGACUGGCAAAAAACGUGGUCAAUAAAAUGUAAAUAUGGCUUGUAUUGCUUUAAGCCAUAACUACAUUU-UUUCUCUCUGUUA- .((((((..(((.((((((......--)))))).)).)..))).))).((((.((.....((((((((..(((((((((........)))))))))..)))))))-)....)).)))).- ( -28.80) >DroEre_CAF1 66169 113 - 1 AUUUCCAGCCGCAUUGUGUAUUGAC--AUACAAGGCAGACUGGCAAAAAACAUGGUCAAUAAAAGUUAAAUAUGGCUUGUAUUGCUUUAAGCCAUAGCUACAUUU-UUUCUCCCUG---- .((((((((.((.((((((......--)))))).)).).)))).)))...((.((.....((((((((..(((((((((........)))))))))..)).))))-))...)).))---- ( -27.10) >DroYak_CAF1 67167 118 - 1 AUUUCCAACCGCAUUGUGUAUUUGCCAACACAAGGCAGACUGGCAAAAAACGUGGUCAAUAAAAUGUAAAUAUGGCUUGUAUUGCUUUAAGCCAAAACUACAUAUUUUUUUCUCUGUG-- .......(((((.((.(((.((((((.......))))))...)))...)).))))).......(((((....(((((((........)))))))....)))))...............-- ( -26.90) >consensus AUUUCCAACCGCAUUGUGUAUUGAC__AUACAAGGCAGACUGGCAAAAAACGUGGUCAAUAAAAUGUAAAUAUGGCUUGUAUUGCUUUAAGCCAUAACUACAUUU_UUUCUCUCUGUU__ .((((((..(((.((((((........)))))).)).)..))).))).((((.((......(((((((..(((((((((........)))))))))..)))))))......)).)))).. (-24.26 = -25.18 + 0.92)

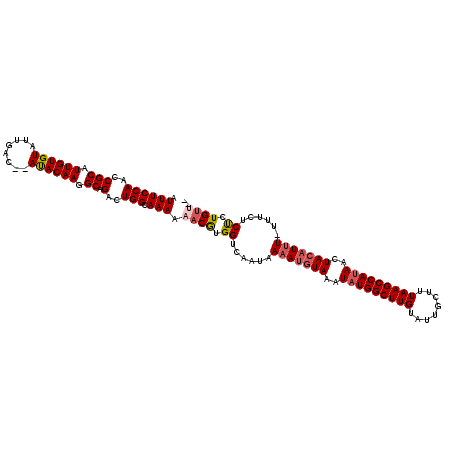
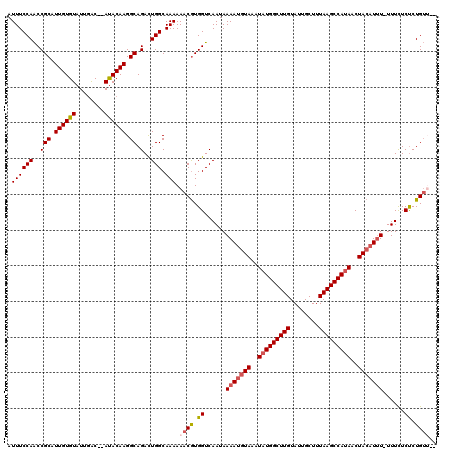
| Location | 10,538,371 – 10,538,489 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10538371 118 + 22224390 ACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAAUCUGCCUUGUAU--GUCAAUACACAAUGCGGUUGGAAAUAAGUAAUCAAGCUGAUGUGAUAAAUUAAAAUUAAACAAUC ...........(((((((...((((..((....(((.((((.((((..(((((--....)))))....)))))))))))...))..))))...))))))).................... ( -22.00) >DroSec_CAF1 64800 118 + 1 ACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAGUCUGCCUUGUAU--GUCAAUACACAAUGCGGUUGGAAAUAAGUAAUCAAGCUGAUGUGAUAAAUUAAAAAUAAACAAAC ...........(((((((...((((..((....(((.((((.((((..(((((--....)))))....)))))))))))...))..))))...))))))).................... ( -22.00) >DroSim_CAF1 68618 118 + 1 ACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAGUCUGCCUUGUAU--GUCAAUACACAAUGCGGUUGGAAAUAAGUAAUCAAGCUGAUGUGAUAAAUUAAAAACAAACAAAC ...........(((((((...((((..((....(((.((((.((((..(((((--....)))))....)))))))))))...))..))))...))))))).................... ( -22.00) >DroEre_CAF1 66204 118 + 1 ACAAGCCAUAUUUAACUUUUAUUGACCAUGUUUUUUGCCAGUCUGCCUUGUAU--GUCAAUACACAAUGCGGCUGGAAAUAAGUAAUCAAGUUGAUGUGAUAAAUUAAAAUUAACCAAAC ......((((((.(((((..((((....(((((....((((.((((..(((((--....)))))....)))))))))))))..)))).)))))))))))..................... ( -20.50) >DroYak_CAF1 67205 120 + 1 ACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAGUCUGCCUUGUGUUGGCAAAUACACAAUGCGGUUGGAAAUAAGUAAUCAAGUUGAUGUGAUAAAUUAAAAUUAAACAAAC ...........(((((((...((((..((....(((.((((.((((.((((((........)))))).)))))))))))...))..))))...))))))).................... ( -26.80) >consensus ACAAGCCAUAUUUACAUUUUAUUGACCACGUUUUUUGCCAGUCUGCCUUGUAU__GUCAAUACACAAUGCGGUUGGAAAUAAGUAAUCAAGCUGAUGUGAUAAAUUAAAAUUAAACAAAC ...........(((((((...((((..((....(((.((((.((((.((((............)))).)))))))))))...))..))))...))))))).................... (-20.26 = -20.18 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:24 2006