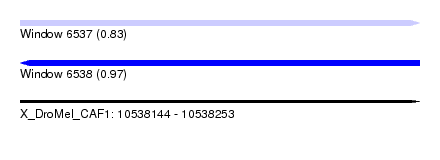
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,538,144 – 10,538,253 |
| Length | 109 |
| Max. P | 0.970225 |

| Location | 10,538,144 – 10,538,253 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.98 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10538144 109 + 22224390 CAUGUGCUUCAACUGAAGCUGUAGCUGAGUUCUCGUUUCUGCAGCUGCAGCACCACUGAUAAUGGACUGACAGCGAAAGCAAAACCAACCACCCACUCAAC---CAAACCAA ....(((((...(((..((((((((((((.........)).)))))))))).(((.......))).....)))...)))))....................---........ ( -27.20) >DroSec_CAF1 64588 109 + 1 CAUGUGCUUCAGCUGAAGCUGAAGCUGAGUUCUCGUUUCUGCAGCUGCAGCACCACUGAUAAUGGACUGACAGUGAAAGCAAAACCAACCACCCACUCAAC---CAAACCAA .....((((((((....))))))))(((((....((((.(((.(((((((..(((.......))).))).))))....))))))).........)))))..---........ ( -28.62) >DroSim_CAF1 68402 109 + 1 CAUGUGCUUCAGCUGAAGCUGAAGCUGAGUUCUCGUUUCUGCAGCUGCAGCACCACUGAUAAUGGACUGACAGCGAAAGCAAAACCAACCACCCACUCAAC---CAAACCAA .....((((((((....))))))))(((((....((((.(((.(((((((..(((.......))).))).))))....))))))).........)))))..---........ ( -30.22) >DroYak_CAF1 66999 105 + 1 CAUGUGCUUCAGCUGAA------GCUGAGUUCCCGUUUCUGCAGCUGCAGCACCACUGAUAAUGGACUGACAGCGAAAGCAAAACCAACCACCCACCC-ACAAACCACCCAA ..((((.((((((....------)))))).....((((.(((.(((((((..(((.......))).))).))))....)))))))............)-))).......... ( -24.40) >consensus CAUGUGCUUCAGCUGAAGCUGAAGCUGAGUUCUCGUUUCUGCAGCUGCAGCACCACUGAUAAUGGACUGACAGCGAAAGCAAAACCAACCACCCACUCAAC___CAAACCAA ....(((((..((((..((((.(((((((.........)).))))).)))).(((.......))).....))))..)))))............................... (-21.60 = -22.98 + 1.37)

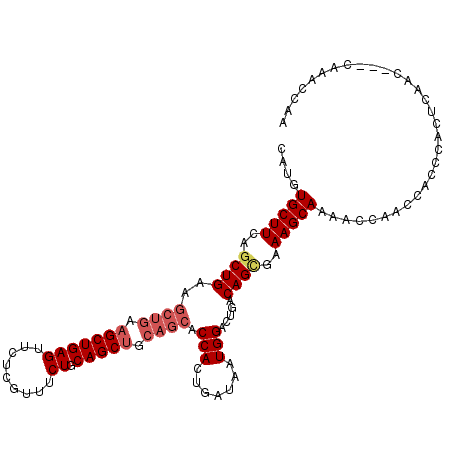
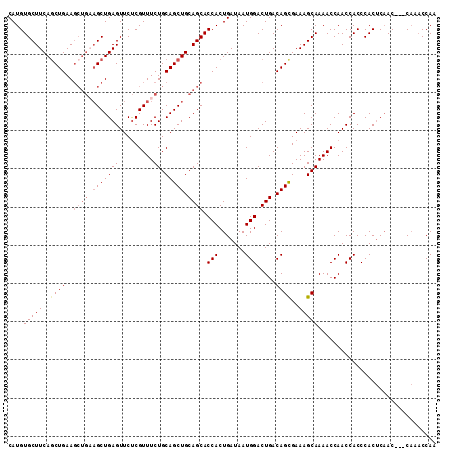
| Location | 10,538,144 – 10,538,253 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -34.44 |
| Energy contribution | -36.12 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10538144 109 - 22224390 UUGGUUUG---GUUGAGUGGGUGGUUGGUUUUGCUUUCGCUGUCAGUCCAUUAUCAGUGGUGCUGCAGCUGCAGAAACGAGAACUCAGCUACAGCUUCAGUUGAAGCACAUG ......((---(((((((...((.....((((((....((((.((((((((.....)))).)))))))).)))))).))...)))))))))..(((((....)))))..... ( -38.90) >DroSec_CAF1 64588 109 - 1 UUGGUUUG---GUUGAGUGGGUGGUUGGUUUUGCUUUCACUGUCAGUCCAUUAUCAGUGGUGCUGCAGCUGCAGAAACGAGAACUCAGCUUCAGCUUCAGCUGAAGCACAUG ........---......(((((..(((.((((((.....(((.((((((((.....)))).)))))))..)))))).)))..)))))((((((((....))))))))..... ( -38.70) >DroSim_CAF1 68402 109 - 1 UUGGUUUG---GUUGAGUGGGUGGUUGGUUUUGCUUUCGCUGUCAGUCCAUUAUCAGUGGUGCUGCAGCUGCAGAAACGAGAACUCAGCUUCAGCUUCAGCUGAAGCACAUG ........---......(((((..(((.((((((....((((.((((((((.....)))).)))))))).)))))).)))..)))))((((((((....))))))))..... ( -42.50) >DroYak_CAF1 66999 105 - 1 UUGGGUGGUUUGU-GGGUGGGUGGUUGGUUUUGCUUUCGCUGUCAGUCCAUUAUCAGUGGUGCUGCAGCUGCAGAAACGGGAACUCAGC------UUCAGCUGAAGCACAUG ....(((......-...(((((..(((.((((((....((((.((((((((.....)))).)))))))).)))))).)))..)))))((------(((....))))).))). ( -35.50) >consensus UUGGUUUG___GUUGAGUGGGUGGUUGGUUUUGCUUUCGCUGUCAGUCCAUUAUCAGUGGUGCUGCAGCUGCAGAAACGAGAACUCAGCUUCAGCUUCAGCUGAAGCACAUG .................(((((..(((.((((((....((((.((((((((.....)))).)))))))).)))))).)))..)))))((((((((....))))))))..... (-34.44 = -36.12 + 1.69)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:21 2006