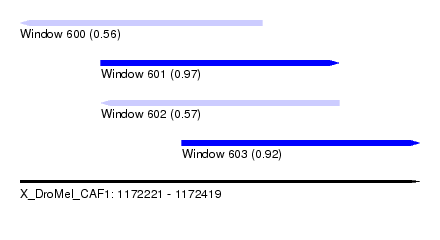
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,172,221 – 1,172,419 |
| Length | 198 |
| Max. P | 0.967328 |

| Location | 1,172,221 – 1,172,341 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.90 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -23.26 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1172221 120 - 22224390 CUUUAAUCGGCCGAUUGAUUAGCAUCUCCAUGCUAAUUAUGGUAUUCCCUUAUCUUUGGCGAUGAUCAGCCGCGGAACUUUUGUUUUUGGUAUGCUGGCCGGCCAAUAGACAGCGAAAAC ......((((((...((((((((((....)))))))))).(((((.((....(((.((((........)))).)))((....))....)).))))))))))).................. ( -32.50) >DroSec_CAF1 8287 116 - 1 CUUUAAUCGGCCGAUUGAUUAGCAUCUCCAUGCUAAUUAUGGUAUUCCCUUAUAUUUGGCAAUGAUCAGCCGCGGAACUCUUGUUUUUGGCCUGCUGG----CCAAUAGACAGCGAAAAC .......((((.(((((((((((((....))))))))))...((((.((........)).))))))).))))......((((((((((((((....))----)))).)))))).)).... ( -33.70) >DroSim_CAF1 9040 116 - 1 CUUUAAUCGGCCGAUUGAUUAGCAUCUCCAUGCUAAUUAUGGUAUUCCCUUAUCUUUGGCGAUGAUCAGCCGCGGAACUUUUGUUUUUGGCCUGCUGG----CCAAUAGACAGCGAAAAC ......(((((((..((((((((((....)))))))))))))).........(((.((((........)))).)))....((((((((((((....))----)))).))))))))).... ( -36.30) >DroEre_CAF1 7395 116 - 1 CUUUAAGCGGCCGAUUGAUUAGCAUCUUCAUGCUAAUUAUGGUAUUUCCCUAUCUUUGGCGAUGAUCAGCCGCGGAACUUUUGUUUUUAGUCUGCUAG----CCACUAAAUAGCGAAAAC ......(((((....((((((((((....))))))))))((((...((((.......)).))..))))))))).....(((((((((((((..(....----).)))))).))))))).. ( -31.20) >DroYak_CAF1 10305 115 - 1 CUUUAAUCGGCCGAUUGAUUAGCAUCUCCGCGCUAAUUAUGGUAUUUCCCUAUCUCUGGCGAUGAUCAGCCGCGAAACUUUUGUU-UUAGUCUGCUUG----CCAAUAAAUAGCGAAAAC .........((((..((((((((........)))))))))))).((((.((((...((((((....(((.(..(((((....)))-)).).))).)))----)))....)))).)))).. ( -27.30) >consensus CUUUAAUCGGCCGAUUGAUUAGCAUCUCCAUGCUAAUUAUGGUAUUCCCUUAUCUUUGGCGAUGAUCAGCCGCGGAACUUUUGUUUUUGGUCUGCUGG____CCAAUAGACAGCGAAAAC ........((((((.((((((((((....)))))))))).............(((.((((........)))).)))..........))))))(((((.............)))))..... (-23.26 = -24.02 + 0.76)

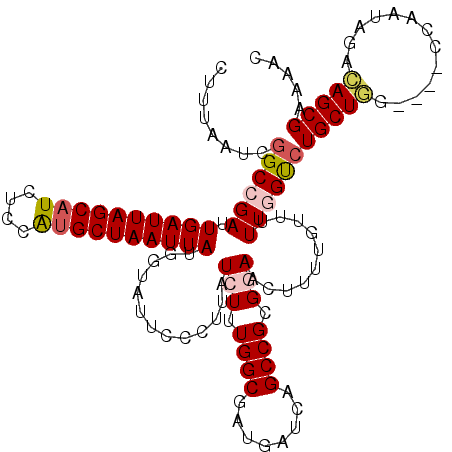
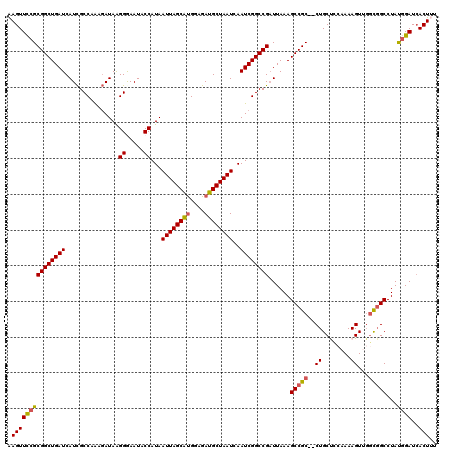
| Location | 1,172,261 – 1,172,379 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -29.68 |
| Energy contribution | -29.76 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1172261 118 + 22224390 AAGUUCCGCGGCUGAUCAUCGCCAAAGAUAAGGGAAUACCAUAAUUAGCAUGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC--CUGCCUCAAAAGUUGGCCGCCUAUGGAUAACUUU (((((((((((((....((((((........((.....))...((((((((....)))))))).....)).))))...))))))--..(((.........))).......)))..)))). ( -34.80) >DroSec_CAF1 8323 118 + 1 GAGUUCCGCGGCUGAUCAUUGCCAAAUAUAAGGGAAUACCAUAAUUAGCAUGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC--CUGCUCCAAAAGUUGGCGGCCUAUGGAUCACUGU .(((((((((((((((.(((.((........)).)))......((((((((....))))))))..))))))))......(((((--(.(((.....))).))))))...))))..))).. ( -41.20) >DroSim_CAF1 9076 118 + 1 AAGUUCCGCGGCUGAUCAUCGCCAAAGAUAAGGGAAUACCAUAAUUAGCAUGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC--CUGCUCCAAAAGUUGGCGGCCUAUUGAUCACUGU .(((.....(((((((.(((......)))..((.....))...((((((((....))))))))..)))))))((((((.(((((--(.(((.....))).))))))...))))))))).. ( -39.70) >DroEre_CAF1 7431 118 + 1 AAGUUCCGCGGCUGAUCAUCGCCAAAGAUAGGGAAAUACCAUAAUUAGCAUGAAGAUGCUAAUCAAUCGGCCGCUUAAAGCCGCCCCUGCCA-CAAAGUUGCCGGCCUACGGAUCACUU- ..(.((((((((((((.(((......))).((......))...((((((((....))))))))..))))))))).....(((((..((....-...))..).))))....))).)....- ( -35.50) >DroYak_CAF1 10340 116 + 1 AAGUUUCGCGGCUGAUCAUCGCCAGAGAUAGGGAAAUACCAUAAUUAGCGCGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC--CUGCUC-AAAAGUCUGUGGCCUACGGAUCACUU- .(((...((((((....((((((.......((......))...(((((((......))))))).....)).))))...))))))--..))).-........(((..(....)..)))..- ( -32.80) >consensus AAGUUCCGCGGCUGAUCAUCGCCAAAGAUAAGGGAAUACCAUAAUUAGCAUGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC__CUGCUCCAAAAGUUGGCGGCCUAUGGAUCACUUU .(((((((((((((((...............((.....))...((((((((....))))))))..))))))))......(((((..((........))...)))))...))))..))).. (-29.68 = -29.76 + 0.08)

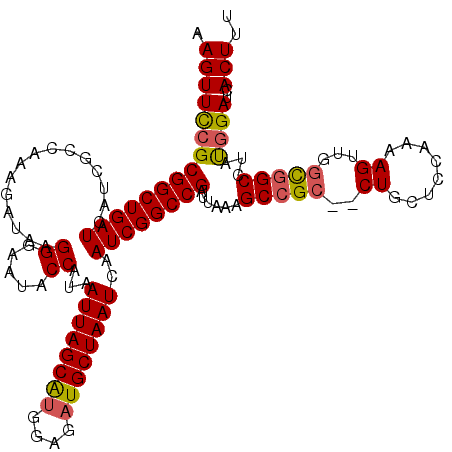
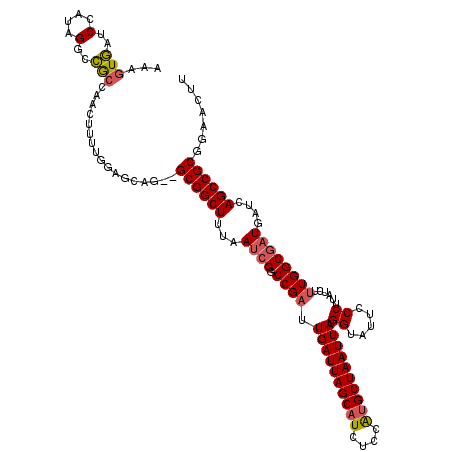
| Location | 1,172,261 – 1,172,379 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -27.54 |
| Energy contribution | -27.90 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1172261 118 - 22224390 AAAGUUAUCCAUAGGCGGCCAACUUUUGAGGCAG--GCGGCUUUAAUCGGCCGAUUGAUUAGCAUCUCCAUGCUAAUUAUGGUAUUCCCUUAUCUUUGGCGAUGAUCAGCCGCGGAACUU .((((..(((.......(((.........)))..--((((((...((((.((((.((((((((((....)))))))))).((((......)))).))))))))....))))))))))))) ( -38.50) >DroSec_CAF1 8323 118 - 1 ACAGUGAUCCAUAGGCCGCCAACUUUUGGAGCAG--GCGGCUUUAAUCGGCCGAUUGAUUAGCAUCUCCAUGCUAAUUAUGGUAUUCCCUUAUAUUUGGCAAUGAUCAGCCGCGGAACUC .......(((..((((((((..(((...)))..)--)))))))....((((.(((((((((((((....))))))))))...((((.((........)).))))))).)))).))).... ( -37.40) >DroSim_CAF1 9076 118 - 1 ACAGUGAUCAAUAGGCCGCCAACUUUUGGAGCAG--GCGGCUUUAAUCGGCCGAUUGAUUAGCAUCUCCAUGCUAAUUAUGGUAUUCCCUUAUCUUUGGCGAUGAUCAGCCGCGGAACUU ....((((((..((((((((..(((...)))..)--)))))))......(((((.((((((((((....)))))))))).((((......)))).)))))..))))))............ ( -38.00) >DroEre_CAF1 7431 118 - 1 -AAGUGAUCCGUAGGCCGGCAACUUUG-UGGCAGGGGCGGCUUUAAGCGGCCGAUUGAUUAGCAUCUUCAUGCUAAUUAUGGUAUUUCCCUAUCUUUGGCGAUGAUCAGCCGCGGAACUU -((((..(((((.(((((((..(((.(-(.((....)).))...)))..))))..((((((((((....))))))))))((((...((((.......)).))..)))))))))))))))) ( -40.80) >DroYak_CAF1 10340 116 - 1 -AAGUGAUCCGUAGGCCACAGACUUUU-GAGCAG--GCGGCUUUAAUCGGCCGAUUGAUUAGCAUCUCCGCGCUAAUUAUGGUAUUUCCCUAUCUCUGGCGAUGAUCAGCCGCGAAACUU -..(((..(....)..)))........-......--((((((...((((((((..((((((((........)))))))))))).....((.......))))))....))))))....... ( -30.40) >consensus AAAGUGAUCCAUAGGCCGCCAACUUUUGGAGCAG__GCGGCUUUAAUCGGCCGAUUGAUUAGCAUCUCCAUGCUAAUUAUGGUAUUCCCUUAUCUUUGGCGAUGAUCAGCCGCGGAACUU ...(((..(....)..))).................((((((...((((.((((.((((((((((....)))))))))).((.....))......))))))))....))))))....... (-27.54 = -27.90 + 0.36)

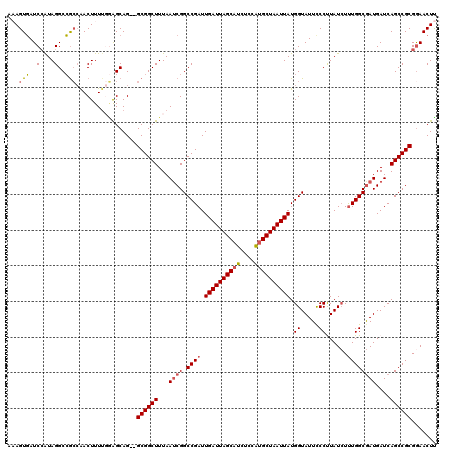
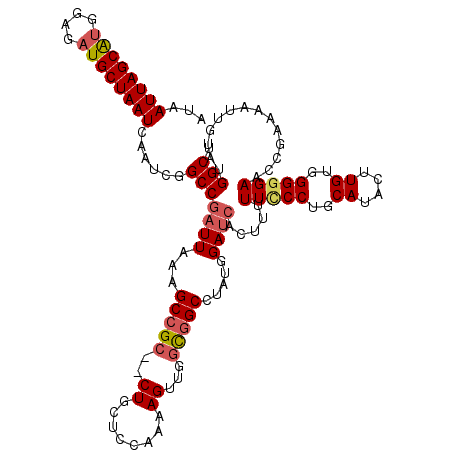
| Location | 1,172,301 – 1,172,419 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -28.96 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1172301 118 + 22224390 AUAAUUAGCAUGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC--CUGCCUCAAAAGUUGGCCGCCUAUGGAUAACUUUUUCCCUGCAUACUUGUGGGGGAGCCGAAAAUUGUAUGGCU ...((((((((....))))))))....((((........)))).--..(((.(((...(((((..((...))........(((((..((....))..))))))))))...)))...))). ( -35.90) >DroSec_CAF1 8363 118 + 1 AUAAUUAGCAUGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC--CUGCUCCAAAAGUUGGCGGCCUAUGGAUCACUGUUUCCCUGCAUACUUGUGGGGGAACCGAAAAUUUUAUGGCU ...((((((((....)))))))).....((((((((...(((((--(.(((.....))).)))))).....)))).(.(((((((..((....))..))))))).)..........)))) ( -46.20) >DroSim_CAF1 9116 118 + 1 AUAAUUAGCAUGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC--CUGCUCCAAAAGUUGGCGGCCUAUUGAUCACUGUUUCCCUGCAUACUUGUGGGGGAACCGAAAAUUGUAUGGCU ...((((((((....)))))))).....((((((((((.(((((--(.(((.....))).))))))...)))))).(.(((((((..((....))..))))))).)..........)))) ( -47.60) >DroEre_CAF1 7471 118 + 1 AUAAUUAGCAUGAAGAUGCUAAUCAAUCGGCCGCUUAAAGCCGCCCCUGCCA-CAAAGUUGCCGGCCUACGGAUCACUU-UUUCCUGCAUACUUGUGGGGGAACAGAAAAUUGUGUGGCA ...((((((((....)))))))).....(((.((.....)).)))...((((-((.((((.(((.....)))....(((-(..((..((....))..))..)).))..)))).)))))). ( -40.10) >DroYak_CAF1 10380 116 + 1 AUAAUUAGCGCGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC--CUGCUC-AAAAGUCUGUGGCCUACGGAUCACUU-UUCCCUGCAUACUUGUGGGCGAACGGAAAAUUGUGUGGCA ...(((((((......))))))).....(((........)))((--(.((.(-((...(((((.(((((((((......-.)))..........))))))..)))))...))).))))). ( -33.20) >consensus AUAAUUAGCAUGGAGAUGCUAAUCAAUCGGCCGAUUAAAGCCGC__CUGCUCCAAAAGUUGGCGGCCUAUGGAUCACUUUUUCCCUGCAUACUUGUGGGGGAACCGAAAAUUGUAUGGCU ...((((((((....))))))))......(((((((...(((((..((........))...))))).....)))).....(((((..((....))..)))))..............))). (-28.96 = -29.72 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:31 2006