
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,525,935 – 10,526,069 |
| Length | 134 |
| Max. P | 0.832445 |

| Location | 10,525,935 – 10,526,033 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -7.40 |
| Energy contribution | -7.88 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10525935 98 + 22224390 CAGUGGCAAAUGCAAUUUGCGAAAUUAAAACCGAAAGUAGUAGUGAAAACUUUUCACUGACCACCACCGC---------------------CAAACCAAACUCCUUU-CGGCCAUUGUCA (((((((...(((.....)))..........((((((.(((((((((.....))))))(.....).....---------------------........))).))))-)))))))))... ( -24.00) >DroSec_CAF1 57113 98 + 1 CAUUGGCAAAUGCAAUUUGCGAAAUUAAAACCGAAAGUGGUAGUGAAAACUUUUCACCGACCACCAACGC---------------------CAAACCAAACUACUUU-CGACCAUUGUCA ..(((((...(((.....)))...............(((((.(((((.....)))))..)))))....))---------------------))).............-.(((....))). ( -21.80) >DroSim_CAF1 60743 98 + 1 CAUUGGCAAAUGCAAUUUGCGAAAUUAAAACCGAAAGUAGUAGUGAAAACUUUUCACCGACCACCAACGC---------------------CAAACCAAACUCCUUU-CGACCAUUGUCA ...((((((.(((.....)))..........((((((.(((.(((((.....)))))((........)).---------------------........))).))))-))....)))))) ( -17.50) >DroEre_CAF1 59107 90 + 1 CACGGGCGAAUGCAAUUUGCGAAAUUAAAACCGAAAGUAGUAGUGACAACUUUUCACCCACCGCCAC-----------------------------CAAACUCCAUU-CGACCAUUGUCA ....((((..(((.....)))...........((((((.((....)).)))))).......))))..-----------------------------...........-.(((....))). ( -14.40) >DroAna_CAF1 61921 114 + 1 CAUUGGCGAAUGCAAUUUGCGAAAUUAAAACCGAAAGUAGUGG---GAUC-CUUCAGGGACUC-CUCCACUUUGGCCUCCUUUUUGGCCAACUU-CUCAUCUCCAUCCUGGCCAUUGUCA ((.((((....((.....))(((..........(((((.(.((---..((-(.....)))..)-).).)))))((((........))))...))-)..............)))).))... ( -29.40) >consensus CAUUGGCAAAUGCAAUUUGCGAAAUUAAAACCGAAAGUAGUAGUGAAAACUUUUCACCGACCACCACCGC_____________________CAAACCAAACUCCUUU_CGACCAUUGUCA ....(((((.(((.((((.((..........)).)))).)))(((((.....))))).........................................................))))). ( -7.40 = -7.88 + 0.48)



| Location | 10,525,935 – 10,526,033 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -13.16 |
| Energy contribution | -14.96 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10525935 98 - 22224390 UGACAAUGGCCG-AAAGGAGUUUGGUUUG---------------------GCGGUGGUGGUCAGUGAAAAGUUUUCACUACUACUUUCGGUUUUAAUUUCGCAAAUUGCAUUUGCCACUG ...(((...((.-...))...)))...((---------------------((((.((((((.((((((.....)))))))))))).))............((.....))....))))... ( -27.80) >DroSec_CAF1 57113 98 - 1 UGACAAUGGUCG-AAAGUAGUUUGGUUUG---------------------GCGUUGGUGGUCGGUGAAAAGUUUUCACUACCACUUUCGGUUUUAAUUUCGCAAAUUGCAUUUGCCAAUG ...((.(((.((-((.(((((((((((.(---------------------.((..((((((.((((((.....))))))))))))..)).)...)))....)))))))).))))))).)) ( -29.90) >DroSim_CAF1 60743 98 - 1 UGACAAUGGUCG-AAAGGAGUUUGGUUUG---------------------GCGUUGGUGGUCGGUGAAAAGUUUUCACUACUACUUUCGGUUUUAAUUUCGCAAAUUGCAUUUGCCAAUG ...(((...((.-...))...)))..(((---------------------(((..((((((.((((((.....))))))))))))..)............((.....))....))))).. ( -25.90) >DroEre_CAF1 59107 90 - 1 UGACAAUGGUCG-AAUGGAGUUUG-----------------------------GUGGCGGUGGGUGAAAAGUUGUCACUACUACUUUCGGUUUUAAUUUCGCAAAUUGCAUUCGCCCGUG .....((((.((-((((.((((((-----------------------------((((.((..(.(....).)..)).))))......(((........))))))))).)))))).)))). ( -27.40) >DroAna_CAF1 61921 114 - 1 UGACAAUGGCCAGGAUGGAGAUGAG-AAGUUGGCCAAAAAGGAGGCCAAAGUGGAG-GAGUCCCUGAAG-GAUC---CCACUACUUUCGGUUUUAAUUUCGCAAAUUGCAUUCGCCAAUG ...((.((((..(((((.((.((.(-(((((((((........))))(((((((.(-(..(((.....)-))..---)).))))))).......)))))).))..)).))))))))).)) ( -36.10) >consensus UGACAAUGGUCG_AAAGGAGUUUGGUUUG_____________________GCGGUGGUGGUCGGUGAAAAGUUUUCACUACUACUUUCGGUUUUAAUUUCGCAAAUUGCAUUUGCCAAUG ...((.((((.........................................((..((((((.((((((.....))))))))))))..))...........((.....))....)))).)) (-13.16 = -14.96 + 1.80)

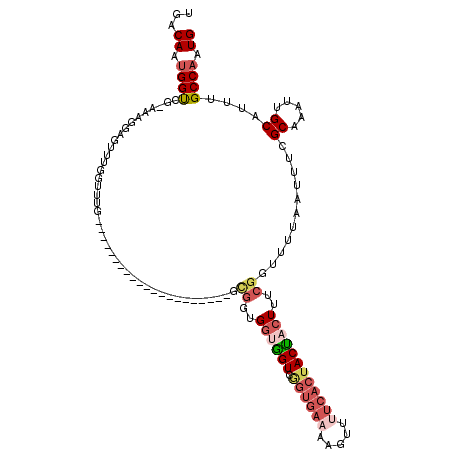
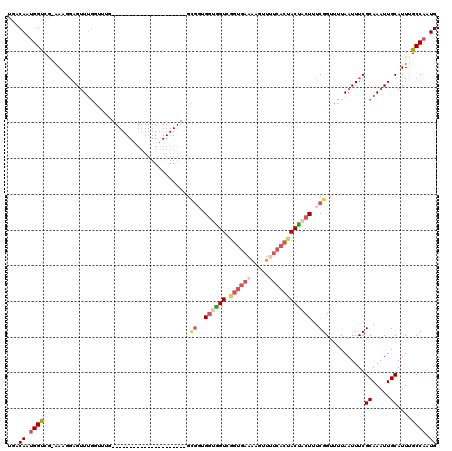
| Location | 10,525,975 – 10,526,069 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -14.65 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10525975 94 + 22224390 UAGUGAAAACUUUUCACUGACCACCACCGCCAAACCAAACUCCUUUCGGCCAUUGUCAGUCGUCUUAUCGGCGACAGCAAUCAUUUCUGCGGCU (((((((.....))))))).........(((................(((....))).((((((.....)))))).(((........)))))). ( -23.10) >DroSec_CAF1 57153 94 + 1 UAGUGAAAACUUUUCACCGACCACCAACGCCAAACCAAACUACUUUCGACCAUUGUCAGUCGUCUUAUCGGUGACAGCAAUCAUUUCUGCGGCU ..(((((.....)))))...........(((......................(((((.(((......))))))))(((........)))))). ( -18.10) >DroSim_CAF1 60783 94 + 1 UAGUGAAAACUUUUCACCGACCACCAACGCCAAACCAAACUCCUUUCGACCAUUGUCAGUCGUCUUAUCGGUGACAGCAAUCAUUUCUGCGGCU ..(((((.....)))))...........(((......................(((((.(((......))))))))(((........)))))). ( -18.10) >DroEre_CAF1 59147 86 + 1 UAGUGACAACUUUUCACCCACCGCCAC--------CAAACUCCAUUCGACCAUUGUCAGUCGCCUUAUCGGUGGCAGCCAUCAUUUCUGCGGCC ..((((.......))))...((((...--------............(((....))).((((((.....)))))).............)))).. ( -18.90) >consensus UAGUGAAAACUUUUCACCGACCACCAACGCCAAACCAAACUCCUUUCGACCAUUGUCAGUCGUCUUAUCGGUGACAGCAAUCAUUUCUGCGGCU ..(((((.....)))))...........(((................(((....))).((((((.....)))))).(((........)))))). (-14.65 = -15.15 + 0.50)


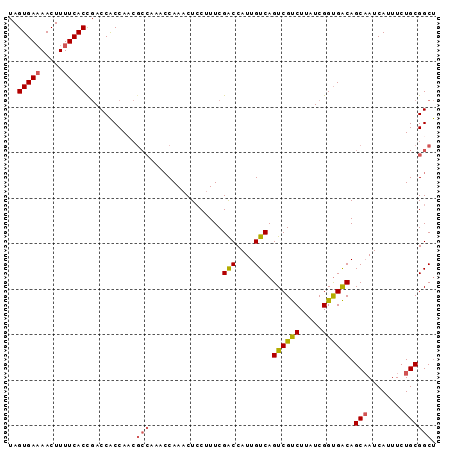
| Location | 10,525,975 – 10,526,069 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -19.01 |
| Energy contribution | -20.07 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10525975 94 - 22224390 AGCCGCAGAAAUGAUUGCUGUCGCCGAUAAGACGACUGACAAUGGCCGAAAGGAGUUUGGUUUGGCGGUGGUGGUCAGUGAAAAGUUUUCACUA ............(((..(..(((((((..((((..((......))((....)).))))...)))))))..)..)))((((((.....)))))). ( -32.70) >DroSec_CAF1 57153 94 - 1 AGCCGCAGAAAUGAUUGCUGUCACCGAUAAGACGACUGACAAUGGUCGAAAGUAGUUUGGUUUGGCGUUGGUGGUCGGUGAAAAGUUUUCACUA ...........(((..(((.((((((((..((((.(..((((....(....)....)))..)..)))))....))))))))..)))..)))... ( -26.10) >DroSim_CAF1 60783 94 - 1 AGCCGCAGAAAUGAUUGCUGUCACCGAUAAGACGACUGACAAUGGUCGAAAGGAGUUUGGUUUGGCGUUGGUGGUCGGUGAAAAGUUUUCACUA ...........(((..(((.((((((((..((((.(..((((...((....))...)))..)..)))))....))))))))..)))..)))... ( -27.60) >DroEre_CAF1 59147 86 - 1 GGCCGCAGAAAUGAUGGCUGCCACCGAUAAGGCGACUGACAAUGGUCGAAUGGAGUUUG--------GUGGCGGUGGGUGAAAAGUUGUCACUA (((((((....)).)))))((((((((..(..(((((......)))))..).....)))--------)))))....(((((.......))))). ( -32.80) >consensus AGCCGCAGAAAUGAUUGCUGUCACCGAUAAGACGACUGACAAUGGUCGAAAGGAGUUUGGUUUGGCGGUGGUGGUCGGUGAAAAGUUUUCACUA ...........(((..(((..(((((((...((.((((.(((...((....))...)))......)))).)).)))))))...)))..)))... (-19.01 = -20.07 + 1.06)

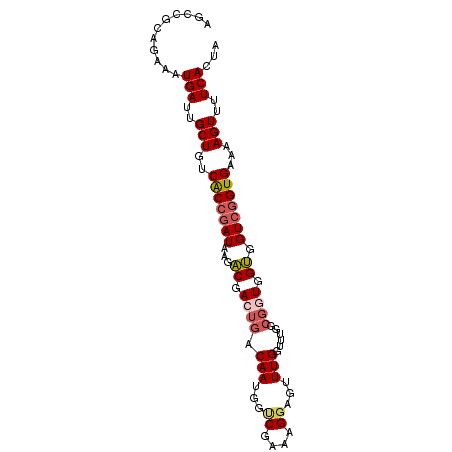

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:11 2006