
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,521,298 – 10,521,435 |
| Length | 137 |
| Max. P | 0.999775 |

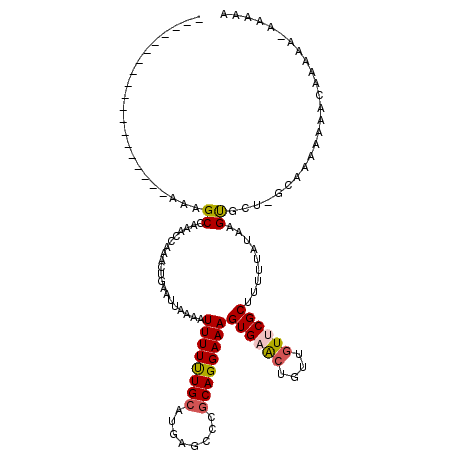
| Location | 10,521,298 – 10,521,399 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.86 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -10.20 |
| Energy contribution | -10.52 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.47 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10521298 101 + 22224390 -------------------AAAGCCAAACCAAGCUGAAUUAAAAUUUUUUGCAUGAGCCCCCAGGAAAGUGAACUGUUGUUCGCUUUUUAUAAGUGCUCGCAAAAAACCAAAAAAAAAAA -------------------..(((........))).........((((((((..(((((....(((((((((((....)))))))))))....).))))))))))))............. ( -24.80) >DroSec_CAF1 52696 96 + 1 -------------------AAAGCCAAACCAAACUGAAUUAAAAUUUUUUGCAUGAGCCCGCAGGAAAGUGAACUGUUGUUCGCUUUUUAUAAGCAA---CAAAAAAACAAAA--AAAAA -------------------...((.........................(((........)))(((((((((((....)))))))))))....))..---.............--..... ( -15.60) >DroSim_CAF1 56283 89 + 1 -------------------AAAGCCAAACCAAACUGAAUUAAAAUUUUUUGCAUGAGCCCGCAGGAAAGUGAACUGUUGUUCGCUUUUUAUAAGUGC----------UCGCAA--AAAAA -------------------.........................((((((((..((((.(...(((((((((((....)))))))))))....).))----------))))))--)))). ( -24.40) >DroEre_CAF1 54706 98 + 1 -------------------AAAGCCAAACCAAACUGAAUUAAAAUUUUCUGCAUGAGCCCGCAGGAAAGUGAACAACAGU-CGCUU-UUAUAAGUGCUGGCAAAAAAAAAA-AAAAAAUA -------------------...((((......((((........((((((((........))))))))........))))-(((((-....))))).))))..........-........ ( -20.89) >DroYak_CAF1 54371 112 + 1 AAGAAAAAACCAAAAGAAAAAAGCCAAACCAAAGUGAAUUAAAAUUUUUUGCAUGAGCCCGCAGGAAAGUGAGGAACAGUACGCUU-UUCUAAGUGCUGGCAAAAA-------AGGCAUA ......................(((..((....)).........((((((((...(((.(..(((((((((..........)))))-))))..).))).)))))))-------))))... ( -23.60) >consensus ___________________AAAGCCAAACCAAACUGAAUUAAAAUUUUUUGCAUGAGCCCGCAGGAAAGUGAACUGUUGUUCGCUUUUUAUAAGUGCU_GCAAAAAAACAAAAA_AAAAA ......................((....................((((((((........))))))))((((((....)))))).........))......................... (-10.20 = -10.52 + 0.32)

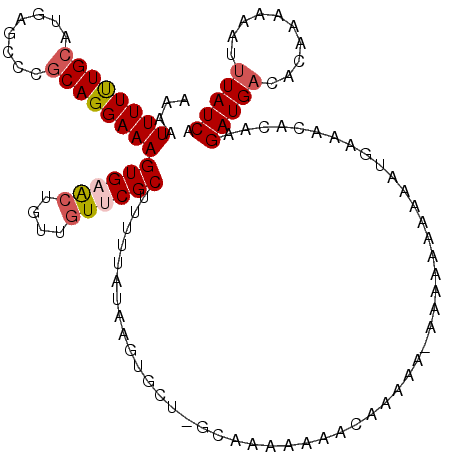
| Location | 10,521,319 – 10,521,435 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -11.34 |
| Energy contribution | -12.02 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10521319 116 + 22224390 AAAAUUUUUUGCAUGAGCCCCCAGGAAAGUGAACUGUUGUUCGCUUUUUAUAAGUGCUCGCAAAAAACCAAAAAAAAAAAAAAAAUGAAACACAAGAUGACACAAAAAAUUUAUCA ....((((((((..(((((....(((((((((((....)))))))))))....).))))))))))))............................(((((..........))))). ( -24.80) >DroSec_CAF1 52717 111 + 1 AAAAUUUUUUGCAUGAGCCCGCAGGAAAGUGAACUGUUGUUCGCUUUUUAUAAGCAA---CAAAAAAACAAAA--AAAAAAAAAAUUAAACACAAGAUGACACAAAAAAUGUAUCA ..((((((((((....))..((.(((((((((((....)))))))))))....))..---.............--....))))))))..........(((.(((.....))).))) ( -17.50) >DroSim_CAF1 56304 104 + 1 AAAAUUUUUUGCAUGAGCCCGCAGGAAAGUGAACUGUUGUUCGCUUUUUAUAAGUGC----------UCGCAA--AAAAAAGAAAUGAAACACAAGAUGACACAAAAAAUUUAUCA ....((((((((..((((.(...(((((((((((....)))))))))))....).))----------))))))--))))................(((((..........))))). ( -25.80) >DroEre_CAF1 54727 113 + 1 AAAAUUUUCUGCAUGAGCCCGCAGGAAAGUGAACAACAGU-CGCUU-UUAUAAGUGCUGGCAAAAAAAAAA-AAAAAAUAACAAAUGAAACACAAGAUGACACAAAAAUGUUAUCA ....((((((((........))))))))(((.....(((.-(((((-....))))))))............-.........(....)...)))..(((((((......))))))). ( -24.10) >DroYak_CAF1 54411 108 + 1 AAAAUUUUUUGCAUGAGCCCGCAGGAAAGUGAGGAACAGUACGCUU-UUCUAAGUGCUGGCAAAAA-------AGGCAUAACAACUGAAACACAAGAUGACAUACAAAAUUUAUCA ....((((((((...(((.(..(((((((((..........)))))-))))..).))).)))))))-------).....................(((((..........))))). ( -21.00) >consensus AAAAUUUUUUGCAUGAGCCCGCAGGAAAGUGAACUGUUGUUCGCUUUUUAUAAGUGCU_GCAAAAAAACAAAAA_AAAAAAAAAAUGAAACACAAGAUGACACAAAAAAUUUAUCA ....((((((((........))))))))((((((....))))))...................................................(((((..........))))). (-11.34 = -12.02 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:03 2006