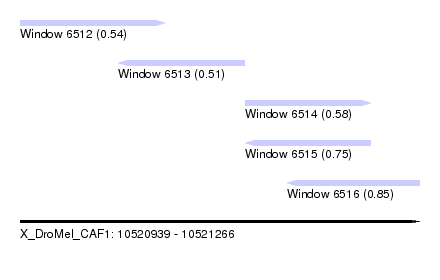
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,520,939 – 10,521,266 |
| Length | 327 |
| Max. P | 0.853222 |

| Location | 10,520,939 – 10,521,058 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10520939 119 + 22224390 CAAGUUUCAAUUGCAAUUGUUGCUGCUAGUUUCUUUACUGCUCUUUUCAGUCAUUAUUUUUUUUUUUUUGUAGCUCGGUGAAAUCAAUGUGUAGGU-UUAUUUUUACUGCACAACUGGCA ..(((..((((....))))..)))((((((...(((((((..((...(((.................))).))..))))))).....(((((((..-.........))))))))))))). ( -24.83) >DroSim_CAF1 55943 100 + 1 CAAGUUUCAAUUGCAAUUGUUGCUGCUGGUUUCUUUACU------------CA-------UUUUUUUUUUUAGCUCUGUGAAAUCAAUGUGUAGGU-UUAUUUUUACUGCACAACUGGCA ..(((..((((....))))..)))((..((...(((((.------------..-------.................))))).....(((((((..-.........)))))))))..)). ( -19.35) >DroEre_CAF1 54358 108 + 1 CAAGUUACAAUUGCAAUUGUUGCUGCUGGUUUCUUUACGGCCCUUUUAAGUCA------------UUUUUCAGAUCGGCGAAAUCAAUGUGUAGCUUUUAUCCUUACUGCACAACUAGCA ..(((.(((((....))))).)))((((((.........(((..(((.((...------------...)).)))..)))........(((((((............))))))))))))). ( -23.00) >DroYak_CAF1 53989 109 + 1 CCAGUUUCAAUUGCAAUUGUGGCUGCUGCUUUCUUUGCUGCUAUUUUGAGUCAG----------UUUUUUUAGCUCGGUGAAAUCAAUGUGUAGAU-UUAUUCUUACUGCACAACUGGCA (((((((((...((((....(((....)))....)))).......(((((..((----------.....))..))))))))).....(((((((..-.........)))))))))))).. ( -25.30) >consensus CAAGUUUCAAUUGCAAUUGUUGCUGCUGGUUUCUUUACUGCUCUUUU_AGUCA___________UUUUUUUAGCUCGGUGAAAUCAAUGUGUAGGU_UUAUUCUUACUGCACAACUGGCA ..(((..((((....))))..)))((((((...(((((.......................................))))).....(((((((............))))))))))))). (-15.53 = -15.53 + 0.00)


| Location | 10,521,019 – 10,521,123 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.77 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -19.97 |
| Energy contribution | -20.65 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10521019 104 - 22224390 ACGGUUAAGUUAACUUGGCCUUGCC------AGCAAUGUGUUGUAUUGGUUAUUGAUUUGGC---------CCUCCUACUUGCCAGUUGUGCAGUAAAAAUAA-ACCUACACAUUGAUUU ..(((((((....))))))).....------..((((((((..((((..((((((..(((((---------..........))))).....)))))).)))).-....)))))))).... ( -27.60) >DroSec_CAF1 52418 104 - 1 ACGGUUAAGUUAACUUGGCCUUGCC------AGCAAUGUGUUGUAUUGGUUAUUGAUUUGGC---------CCUCCUACUUGCCAGUUGUGCAGUAAAAAUAA-ACCUACACAUUGAUUU ..(((((((....))))))).....------..((((((((..((((..((((((..(((((---------..........))))).....)))))).)))).-....)))))))).... ( -27.60) >DroSim_CAF1 56004 104 - 1 ACGGUUAAGUUAACUUGGCCUUGCC------AGCAAUGUGUUGUAUUGGUUAUUGAUUUGGC---------CCUCCUACUUGCCAGUUGUGCAGUAAAAAUAA-ACCUACACAUUGAUUU ..(((((((....))))))).....------..((((((((..((((..((((((..(((((---------..........))))).....)))))).)))).-....)))))))).... ( -27.60) >DroEre_CAF1 54426 101 - 1 ACGGUUAAGCUAACUUGGCCUUG----------CCAAGUGUUGUAUUGGUUAUUGAUUUGGC---------CCUCCUACUUGCUAGUUGUGCAGUAAGGAUAAAAGCUACACAUUGAUUU ..(((((((....)))))))...----------.(((((((.((...(((((......))))---------).(((((((.((.......))))).)))).....)).)))).))).... ( -25.20) >DroYak_CAF1 54059 119 - 1 ACGGUUAAGCUAACUUGGCCUUGUGUUGUUGGCCCAAGUGUUGUAUUGGUUAUUGAUUUGCCUUGCUGCCACCUCCUACUUGCCAGUUGUGCAGUAAGAAUAA-AUCUACACAUUGAUUU ..(((((((....))))))).(((((.......((((........)))).....((((((.((((((((((.((..........)).)).))))))))..)))-))).)))))....... ( -28.80) >consensus ACGGUUAAGUUAACUUGGCCUUGCC______AGCAAUGUGUUGUAUUGGUUAUUGAUUUGGC_________CCUCCUACUUGCCAGUUGUGCAGUAAAAAUAA_ACCUACACAUUGAUUU ..(((((((....))))))).............((((((((..((((..((((((..(((((...................))))).....)))))).))))......)))))))).... (-19.97 = -20.65 + 0.68)


| Location | 10,521,123 – 10,521,226 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -24.74 |
| Energy contribution | -25.06 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10521123 103 + 22224390 UGCCCUGCGAACAGUU------GCCAAGCUGAAAAUGGACAGGUGGCGCGGGCAGGGGGAUUCCCCG----------GGGGCAAUAAAUUAACAGCCCGAUGGCAAGAGCAAAG-AAAAU (((((.(((..((.((------((((.........))).))).)).))))))))((((....)))).----------.((((............))))....((....))....-..... ( -34.10) >DroSec_CAF1 52522 102 + 1 UGCCCUGCGAACAGUU------GCCAAGCUGAAAAUGGACAGGUGGCGCGGGCAGG-GGAUUCCCCG----------GGGGCAAUAAAUUAACAGCCCGAUGGCGAGAGCAAAG-AAAAU (((((.(((..((.((------((((.........))).))).)).))))))))((-((...)))).----------.((((............))))....((....))....-..... ( -35.50) >DroSim_CAF1 56108 102 + 1 UGCCCUGCGAACAGUU------GCCAAGCUGAAAAUGGACAGGUGGCGCGGGCAGG-GGAUUCCCCG----------GGGGCAAUAAAUUAACAGCCCGAUGGCGAGAGCAAAG-AAAAU (((((.(((..((.((------((((.........))).))).)).))))))))((-((...)))).----------.((((............))))....((....))....-..... ( -35.50) >DroEre_CAF1 54527 108 + 1 UGCCCUGCGAACAGUUGCCGUUGCCAAGCUGAAAAUGGACAGGUGGUGCUGGAGG--GGAUUCCCCAG---------GGGGCCAUAAAUUAACAGCCCGACGGCGAGAGCGAAAGAAAA- .((.(((....)))(((((((((....((((..(((......(((((.((...((--((...))))..---------)).)))))..)))..)))).)))))))))..)).........- ( -33.60) >DroYak_CAF1 54178 114 + 1 UGCCCUGCGAACAGUU------GCCAGGCUGAAAAUGGACAGGUGGCGCUGGAGGGGGGAUUCCCCGGGGAAGGGGGGGGGCAAUAAAUUAACAGCCCGAUGGCGAGAGCAAAAGAAAAU ..((((.(...((((.------((((..(((........))).))))))))..).)))).((((((......))))))((((............))))....((....)).......... ( -40.20) >consensus UGCCCUGCGAACAGUU______GCCAAGCUGAAAAUGGACAGGUGGCGCGGGCAGG_GGAUUCCCCG__________GGGGCAAUAAAUUAACAGCCCGAUGGCGAGAGCAAAG_AAAAU ((((.(((..............((((..(((........))).)))).(((((.(...((((((((...........)))).....))))..).)))))...))).).)))......... (-24.74 = -25.06 + 0.32)


| Location | 10,521,123 – 10,521,226 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -23.21 |
| Energy contribution | -23.41 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10521123 103 - 22224390 AUUUU-CUUUGCUCUUGCCAUCGGGCUGUUAAUUUAUUGCCCC----------CGGGGAAUCCCCCUGCCCGCGCCACCUGUCCAUUUUCAGCUUGGC------AACUGUUCGCAGGGCA .....-...(((.(((((...(((((.((.........))...----------.((((....)))).))))).((((.(((........)))..))))------........)))))))) ( -31.80) >DroSec_CAF1 52522 102 - 1 AUUUU-CUUUGCUCUCGCCAUCGGGCUGUUAAUUUAUUGCCCC----------CGGGGAAUCC-CCUGCCCGCGCCACCUGUCCAUUUUCAGCUUGGC------AACUGUUCGCAGGGCA .....-...(((((((((....((((.((......)).)))).----------(((((....)-))))...))((((.(((........)))..))))------........).)))))) ( -30.20) >DroSim_CAF1 56108 102 - 1 AUUUU-CUUUGCUCUCGCCAUCGGGCUGUUAAUUUAUUGCCCC----------CGGGGAAUCC-CCUGCCCGCGCCACCUGUCCAUUUUCAGCUUGGC------AACUGUUCGCAGGGCA .....-...(((((((((....((((.((......)).)))).----------(((((....)-))))...))((((.(((........)))..))))------........).)))))) ( -30.20) >DroEre_CAF1 54527 108 - 1 -UUUUCUUUCGCUCUCGCCGUCGGGCUGUUAAUUUAUGGCCCC---------CUGGGGAAUCC--CCUCCAGCACCACCUGUCCAUUUUCAGCUUGGCAACGGCAACUGUUCGCAGGGCA -.........(((...(((((.(((((((......))))))).---------((((((.....--.))))))..(((.(((........)))..)))..)))))..(((....)))))). ( -34.70) >DroYak_CAF1 54178 114 - 1 AUUUUCUUUUGCUCUCGCCAUCGGGCUGUUAAUUUAUUGCCCCCCCCCUUCCCCGGGGAAUCCCCCCUCCAGCGCCACCUGUCCAUUUUCAGCCUGGC------AACUGUUCGCAGGGCA .........((((((.((....((((.((......)).))))............((((.....))))..(((.((((.(((........)))..))))------..)))...)))))))) ( -31.20) >consensus AUUUU_CUUUGCUCUCGCCAUCGGGCUGUUAAUUUAUUGCCCC__________CGGGGAAUCC_CCUGCCCGCGCCACCUGUCCAUUUUCAGCUUGGC______AACUGUUCGCAGGGCA ..........(((((.((...(((((.............((((...........)))).............))((((.(((........)))..))))........)))...))))))). (-23.21 = -23.41 + 0.20)


| Location | 10,521,157 – 10,521,266 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -26.64 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10521157 109 - 22224390 GUCGGAGGGCAUCGAGAUUUCUGCCGACAUUUUCCGUGGCAUUUU-CUUUGCUCUUGCCAUCGGGCUGUUAAUUUAUUGCCCC----------CGGGGAAUCCCCCUGCCCGCGCCACCU ...((.(((((..(.((((((..(((.........((((((....-.........)))))).((((.((......)).)))).----------))))))))).)..)))))...)).... ( -34.92) >DroSec_CAF1 52556 108 - 1 GUCGGGGAGAAUUGAGAUUUUUGCCGACAUUUUUCCUGGCAUUUU-CUUUGCUCUCGCCAUCGGGCUGUUAAUUUAUUGCCCC----------CGGGGAAUCC-CCUGCCCGCGCCACCU (.((((((((...((((....(((((..........)))))...)-)))...))))......((((.((......)).)))).----------(((((....)-)))))))))....... ( -33.50) >DroSim_CAF1 56142 108 - 1 GUCGGAGAGAAUUGAGAUUUUUGCCGACAUUUUCCCUGGCAUUUU-CUUUGCUCUCGCCAUCGGGCUGUUAAUUUAUUGCCCC----------CGGGGAAUCC-CCUGCCCGCGCCACCU ....(((((....((((....(((((..........)))))...)-)))..)))))((....((((.((......)).)))).----------(((((....)-))))...))....... ( -31.50) >DroEre_CAF1 54567 108 - 1 GACGGGGAGAAUUGAGAUUUUUGCCGGCAUUUUUCCUGGC-UUUUCUUUCGCUCUCGCCGUCGGGCUGUUAAUUUAUGGCCCC---------CUGGGGAAUCC--CCUCCAGCACCACCU (((((.((((...((((.....(((((........)))))-...))))....)))).)))))(((((((......))))))).---------((((((.....--.))))))........ ( -43.30) >DroYak_CAF1 54212 120 - 1 GUUGGGGAAAUUUGAGAUUUUUGCCGACAUUUUCCCUGGCAUUUUCUUUUGCUCUCGCCAUCGGGCUGUUAAUUUAUUGCCCCCCCCCUUCCCCGGGGAAUCCCCCCUCCAGCGCCACCU (((((((((....((((....(((((..........)))))...))))..............((((.((......)).))))......))))))((((.....))))...)))....... ( -32.60) >consensus GUCGGGGAGAAUUGAGAUUUUUGCCGACAUUUUCCCUGGCAUUUU_CUUUGCUCUCGCCAUCGGGCUGUUAAUUUAUUGCCCC__________CGGGGAAUCC_CCUGCCCGCGCCACCU (((((.((((((....)))))).)))))...((((((((((........)))..........((((.((......)).))))...........))))))).................... (-26.64 = -26.60 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:01 2006