
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,519,468 – 10,519,600 |
| Length | 132 |
| Max. P | 0.998854 |

| Location | 10,519,468 – 10,519,567 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.26 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -11.89 |
| Energy contribution | -13.42 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.44 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
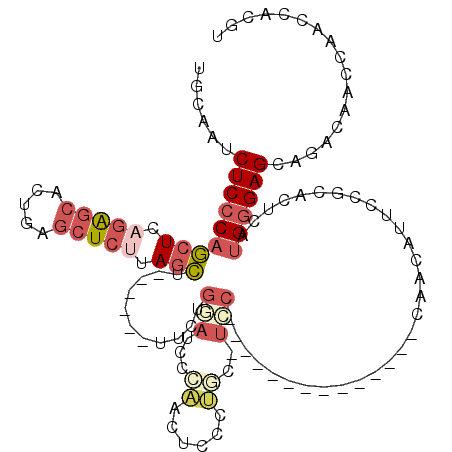
>X_DroMel_CAF1 10519468 99 + 22224390 UGCAAUCUCCCAGCUCAGAGCACUGAGCUCUUAGCU------UUCUGGAUCCCAACUCCCUGC-UCC--------------CAACAUUCCGCACUCAUGGGAGCAGACAACCAACCACGU ((..((((...((((.(((((.....))))).))))------....))))..)).....((((-(((--------------((..............))))))))).............. ( -29.94) >DroSec_CAF1 50977 99 + 1 UGCAGUCUCCCAGCUCAGAGCACUGAGCUCUUAGCU------UUCUGGAUCCCAACUCCCUGC-UCC--------------CAACAUUACGCACUCAUGGGAGCAGACAACCAACCACGU ((..((((...((((.(((((.....))))).))))------....))))..)).....((((-(((--------------((..............))))))))).............. ( -30.24) >DroSim_CAF1 54507 99 + 1 UGCAAUCUCCCAGCUCAGAGCACUGAGCUCUUAGCU------UUCUGGAUCCCAACUCCCUGC-UCC--------------CAACAUUCCGCACUCAUGGGAGCAGACAACCAACCACGU ((..((((...((((.(((((.....))))).))))------....))))..)).....((((-(((--------------((..............))))))))).............. ( -29.94) >DroEre_CAF1 52873 100 + 1 UGCAAUCUCCCUGCUCAGAGCUCUGAGCUCUUAGCU------UUCUGCAUCCAAGCUCCCUGCCUGC--------------CACAUUUCCGCACUUAUGAGAGCAAACAACCAACCACGU ((((........(((.(((((.....))))).))).------...)))).....(((((.((..(((--------------.........)))..)).).))))................ ( -19.90) >DroYak_CAF1 52414 87 + 1 UGCAAUCUCCCAGCUCCGAGCUCCGAGCUCAGAGCU--------------CCAAGCUCUUAG-CUCU--------------UAGCUUUCUGG----AUGGGAGCAGACAAGCAGCCACGU (((...((((((..((((((((..(((((.(((((.--------------....))))).))-))).--------------.)))))...))----))))))).......)))....... ( -34.20) >DroAna_CAF1 56618 110 + 1 UGCAA-CUCCCAUCUCCUGACACCCGAAUCUGAAAUCCUUAAUCCUGUAUCCUGAAUCCUCAA-UUCCGACUUGUGCAGCACACUUUUUAGCACUCGUGGGAGCAGCCAACC-------- .((..-((((((((....))....(((..((((((.........((((((.(.((((.....)-))).)....))))))......))))))...)))))))))..)).....-------- ( -19.26) >consensus UGCAAUCUCCCAGCUCAGAGCACUGAGCUCUUAGCU______UUCUGGAUCCCAACUCCCUGC_UCC______________CAACAUUCCGCACUCAUGGGAGCAGACAACCAACCACGU ......(((((((((.(((((.....))))).)))...........(((...((......))..)))..............................))))))................. (-11.89 = -13.42 + 1.53)

| Location | 10,519,468 – 10,519,567 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.26 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
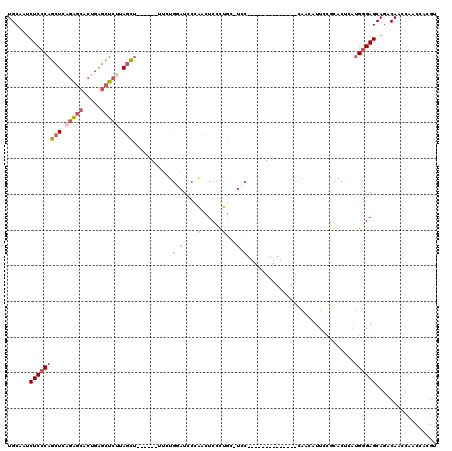
>X_DroMel_CAF1 10519468 99 - 22224390 ACGUGGUUGGUUGUCUGCUCCCAUGAGUGCGGAAUGUUG--------------GGA-GCAGGGAGUUGGGAUCCAGAA------AGCUAAGAGCUCAGUGCUCUGAGCUGGGAGAUUGCA ..(..(((..((.((((((((((...((.....))..))--------------)))-))))).))......(((....------((((.(((((.....))))).)))).))))))..). ( -40.20) >DroSec_CAF1 50977 99 - 1 ACGUGGUUGGUUGUCUGCUCCCAUGAGUGCGUAAUGUUG--------------GGA-GCAGGGAGUUGGGAUCCAGAA------AGCUAAGAGCUCAGUGCUCUGAGCUGGGAGACUGCA ..(..(((..((.((((((((((...((.....))..))--------------)))-))))).))......(((....------((((.(((((.....))))).)))).))))))..). ( -41.50) >DroSim_CAF1 54507 99 - 1 ACGUGGUUGGUUGUCUGCUCCCAUGAGUGCGGAAUGUUG--------------GGA-GCAGGGAGUUGGGAUCCAGAA------AGCUAAGAGCUCAGUGCUCUGAGCUGGGAGAUUGCA ..(..(((..((.((((((((((...((.....))..))--------------)))-))))).))......(((....------((((.(((((.....))))).)))).))))))..). ( -40.20) >DroEre_CAF1 52873 100 - 1 ACGUGGUUGGUUGUUUGCUCUCAUAAGUGCGGAAAUGUG--------------GCAGGCAGGGAGCUUGGAUGCAGAA------AGCUAAGAGCUCAGAGCUCUGAGCAGGGAGAUUGCA ...........(((...(((((......((.....((((--------------.(((((.....)))))..))))...------.)).....((((((....)))))).)))))...))) ( -29.30) >DroYak_CAF1 52414 87 - 1 ACGUGGCUGCUUGUCUGCUCCCAU----CCAGAAAGCUA--------------AGAG-CUAAGAGCUUGG--------------AGCUCUGAGCUCGGAGCUCGGAGCUGGGAGAUUGCA ...(((..((......))..)))(----((....((((.--------------.(((-((..(((((..(--------------....)..)))))..)))))..)))).)))....... ( -37.50) >DroAna_CAF1 56618 110 - 1 --------GGUUGGCUGCUCCCACGAGUGCUAAAAAGUGUGCUGCACAAGUCGGAA-UUGAGGAUUCAGGAUACAGGAUUAAGGAUUUCAGAUUCGGGUGUCAGGAGAUGGGAG-UUGCA --------.....((.((((((((..(..((....))..).((((((.((((((((-((...(((((........)))))...)))))).))))...))).)))..).))))))-).)). ( -31.60) >consensus ACGUGGUUGGUUGUCUGCUCCCAUGAGUGCGGAAAGUUG______________GGA_GCAGGGAGUUGGGAUCCAGAA______AGCUAAGAGCUCAGUGCUCUGAGCUGGGAGAUUGCA ...........(((...(((((...............................(((..(((....)))...)))..........((((.(((((.....))))).)))))))))...))) (-18.14 = -18.78 + 0.64)


| Location | 10,519,504 – 10,519,600 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -9.96 |
| Energy contribution | -10.80 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
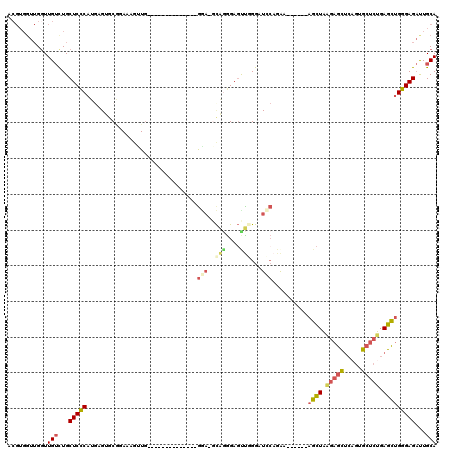
>X_DroMel_CAF1 10519504 96 + 22224390 UUCUGGAUCCCAACUCCCUGC-UCCCAACAUUCCGCACUCAUGGGAGCAGACAACCAACCACGUCACCAAGUCAUCCAGGCAUUCAACCAUAUGC-------CA ...(((((..(.......(((-(((((..............))))))))(((..........))).....)..)))))(((((........))))-------). ( -26.74) >DroSec_CAF1 51013 103 + 1 UUCUGGAUCCCAACUCCCUGC-UCCCAACAUUACGCACUCAUGGGAGCAGACAACCAACCACGUCACCAAGUCAUCCAGGCAUUCAACCAUAUGCCAUGUGCCA ...(((((..(.......(((-(((((..............))))))))(((..........))).....)..)))))(((((........)))))........ ( -26.74) >DroSim_CAF1 54543 103 + 1 UUCUGGAUCCCAACUCCCUGC-UCCCAACAUUCCGCACUCAUGGGAGCAGACAACCAACCACGUCACCAAGUCAUCCAGGCAUUCAACCAUAUGCCAUGUGCCA ...(((((..(.......(((-(((((..............))))))))(((..........))).....)..)))))(((((........)))))........ ( -26.74) >DroEre_CAF1 52909 104 + 1 UUCUGCAUCCAAGCUCCCUGCCUGCCACAUUUCCGCACUUAUGAGAGCAAACAACCAACCACGUCCCCAUGCCAUCCAGGCAUUCAACCAUGUGCCAUGUGCCA ....((((....(((((.((..(((.........)))..)).).))))...........(((((....(((((.....)))))......)))))....)))).. ( -18.60) >DroYak_CAF1 52450 91 + 1 --------CCAAGCUCUUAG-CUCUUAGCUUUCUGG----AUGGGAGCAGACAAGCAGCCACGUCACCAUGCCAUCCAGGCAUUCAACCAUAUGGCAUGUUCCA --------....((((((.(-((((((.((....))----.)))))))....))).)))........((((((((...((.......))..))))))))..... ( -23.40) >consensus UUCUGGAUCCCAACUCCCUGC_UCCCAACAUUCCGCACUCAUGGGAGCAGACAACCAACCACGUCACCAAGUCAUCCAGGCAUUCAACCAUAUGCCAUGUGCCA .................((((.(((((..............)))))))))............................(((((........)))))........ ( -9.96 = -10.80 + 0.84)

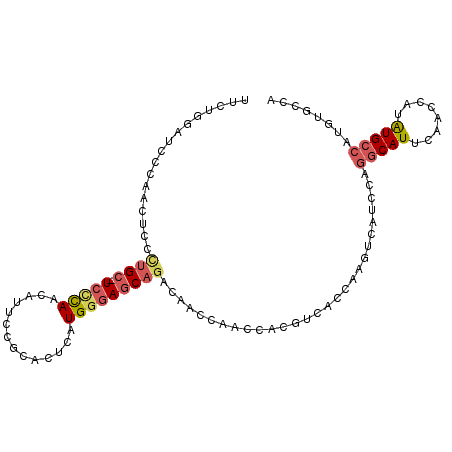

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:57 2006