
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,517,989 – 10,518,149 |
| Length | 160 |
| Max. P | 0.962496 |

| Location | 10,517,989 – 10,518,109 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -27.94 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10517989 120 + 22224390 UAUUCCGUGGCAGUUAAUAUACACUACUGAAAUAUCCAGUUGCAAAGCCGCCUCAUAAACGGUGGCCAUAUGGCUUCCCCAAACGGCGACACUUACAAACGGCACACGUUCGGUGGCCAA ..(((.((((..((......)).)))).))).......(((((...((((((........))))))....(((.....)))....)))))..........(((.(((.....)))))).. ( -31.90) >DroSec_CAF1 49524 120 + 1 UAUUCCGUGGCAGUUGAUAUACACUACUGAGAUAUCCAAUUGCAAAGCCGCCUCAUAAACGGUGGCCAUAUGGUUUCCCCAAACGGCGACACUUACAAAUGGUACACGUUCGGUGGCCAA .........(((((((((((.(.(....).))))).)))))))...((((((.....((((...(((((..(((.((.((....)).)).))).....)))))...)))).))))))... ( -32.40) >DroSim_CAF1 53048 120 + 1 UAUUCCGUGGCAGUUGAUAUACACUACUGAGAUAUCCAAUUGCAAAGCCGCCUCAUAAACGGUGGCCAUAUGGCUUCCCCAAACGGCGACACUUACAAAUGGCACACGUUCGGUGGCCAA ......((((((((((((((.(.(....).))))).))))))).....((((........((.((((....)))).))......)))).))).......((((.(((.....))))))). ( -34.94) >DroEre_CAF1 51470 120 + 1 UAUGCCCUGGCAGUUGAUAUACACUACUGAGAUAUCCAGAUGCAAAGCCGCCUCAUAAACGUUGGCCAUUUGGCUGCCCCAAAUGGCGGCACUUACAAAUGGCACACGUUCGGUGGCCAA ..(((.((((((((...........))))......))))..)))..((((((.....(((((..((((((((..((((((....)).))))....))))))))..))))).))))))... ( -42.50) >DroYak_CAF1 50943 120 + 1 UAUUCCCUGGCAGUUGAUGUAGACCACCGAGAUGUCCAGCUGCAAUGCCGCCUCAUAAACGGUGGCCAUUUGCCUGCCCCAAAUGGCGGCACUUACAAAUGGCACACGUUCGGUGGCCAA .......((((.(((......)))(((((((.((((((...((((.((((((........))))))...)))).((((((....)).))))........))).)))..))))))))))). ( -44.10) >consensus UAUUCCGUGGCAGUUGAUAUACACUACUGAGAUAUCCAGUUGCAAAGCCGCCUCAUAAACGGUGGCCAUAUGGCUUCCCCAAACGGCGACACUUACAAAUGGCACACGUUCGGUGGCCAA .........(((((((((((...(....)..)))).)))))))...((((((.....((((...(((((.((...((.((....)).))......)).)))))...)))).))))))... (-27.94 = -28.10 + 0.16)


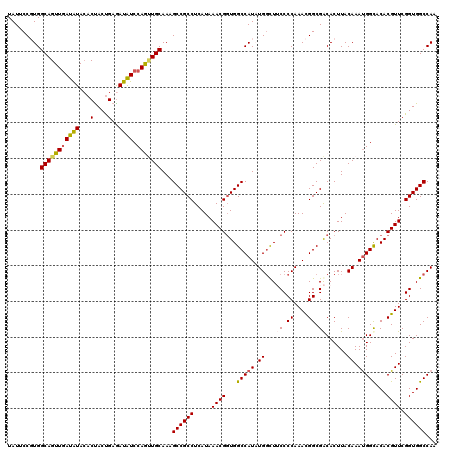
| Location | 10,517,989 – 10,518,109 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -41.34 |
| Consensus MFE | -32.12 |
| Energy contribution | -32.28 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10517989 120 - 22224390 UUGGCCACCGAACGUGUGCCGUUUGUAAGUGUCGCCGUUUGGGGAAGCCAUAUGGCCACCGUUUAUGAGGCGGCUUUGCAACUGGAUAUUUCAGUAGUGUAUAUUAACUGCCACGGAAUA .(((((((((((((.(((.((........)).))))))))))((...))...))))))((((......(((((...((((((((((...))))))..))))......))))))))).... ( -37.60) >DroSec_CAF1 49524 120 - 1 UUGGCCACCGAACGUGUACCAUUUGUAAGUGUCGCCGUUUGGGGAAACCAUAUGGCCACCGUUUAUGAGGCGGCUUUGCAAUUGGAUAUCUCAGUAGUGUAUAUCAACUGCCACGGAAUA .((((((.((((((.((..((((....))))..))))))))((....))...))))))((((...(((((...((........))...)))))(((((........))))).)))).... ( -38.40) >DroSim_CAF1 53048 120 - 1 UUGGCCACCGAACGUGUGCCAUUUGUAAGUGUCGCCGUUUGGGGAAGCCAUAUGGCCACCGUUUAUGAGGCGGCUUUGCAAUUGGAUAUCUCAGUAGUGUAUAUCAACUGCCACGGAAUA ((((...)))).((((..(((.(((((((.((((((....(.((..(((....)))..)).)......))))))))))))).)))........(((((........)))))))))..... ( -38.40) >DroEre_CAF1 51470 120 - 1 UUGGCCACCGAACGUGUGCCAUUUGUAAGUGCCGCCAUUUGGGGCAGCCAAAUGGCCAACGUUUAUGAGGCGGCUUUGCAUCUGGAUAUCUCAGUAGUGUAUAUCAACUGCCAGGGCAUA ...(((...((((((..((((((((....((((.((....))))))..))))))))..))))))....(((((...(((((((((.....))))..)))))......)))))..)))... ( -43.40) >DroYak_CAF1 50943 120 - 1 UUGGCCACCGAACGUGUGCCAUUUGUAAGUGCCGCCAUUUGGGGCAGGCAAAUGGCCACCGUUUAUGAGGCGGCAUUGCAGCUGGACAUCUCGGUGGUCUACAUCAACUGCCAGGGAAUA ..(((((((((..((((.(((.((((((.(((((((((..(.((..(((.....))).)).)..))..))))))))))))).))))))).)))))))))..................... ( -48.90) >consensus UUGGCCACCGAACGUGUGCCAUUUGUAAGUGUCGCCGUUUGGGGAAGCCAUAUGGCCACCGUUUAUGAGGCGGCUUUGCAACUGGAUAUCUCAGUAGUGUAUAUCAACUGCCACGGAAUA .......((((..((((.(((.(((((((.((((((....(.((..(((....)))..)).)......))))))))))))).))))))).)).(((((........)))))...)).... (-32.12 = -32.28 + 0.16)


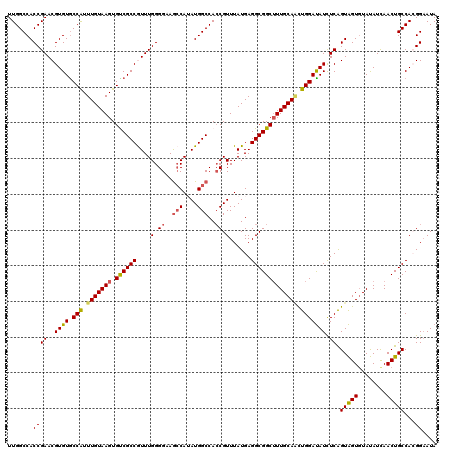
| Location | 10,518,029 – 10,518,149 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -42.98 |
| Consensus MFE | -35.18 |
| Energy contribution | -35.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10518029 120 + 22224390 UGCAAAGCCGCCUCAUAAACGGUGGCCAUAUGGCUUCCCCAAACGGCGACACUUACAAACGGCACACGUUCGGUGGCCAAAAGACGGGGUAAUUCGGUUGCAUUGCGAAAAGUGUGCAAA (((((.((((((........))))))....((..((((((....(....)..........(((.(((.....)))))).......)))).))..)).)))))(((((.......))))). ( -35.50) >DroSec_CAF1 49564 120 + 1 UGCAAAGCCGCCUCAUAAACGGUGGCCAUAUGGUUUCCCCAAACGGCGACACUUACAAAUGGUACACGUUCGGUGGCCAAAAGACGGGGUAAUUCGGUGGCAUUGCGAAAUGUGUGCAAA ......((((((........))))))(((..(((.((.((....)).)).))).....)))(((((((((((...((((.....((((....)))).))))....)).)))))))))... ( -35.60) >DroSim_CAF1 53088 120 + 1 UGCAAAGCCGCCUCAUAAACGGUGGCCAUAUGGCUUCCCCAAACGGCGACACUUACAAAUGGCACACGUUCGGUGGCCAAAAGACGGGGUAAUUCGGUGGCAUUGCGAAAUGUGUGCAAA .((((.((((((...........((((....)))).((((....(....).........((((.(((.....)))))))......))))......)))))).)))).............. ( -40.10) >DroEre_CAF1 51510 120 + 1 UGCAAAGCCGCCUCAUAAACGUUGGCCAUUUGGCUGCCCCAAAUGGCGGCACUUACAAAUGGCACACGUUCGGUGGCCAAAAGGCGGGGUAAUUCGGCGGCAGUGCGGAAAGUGUGCAAA .(((..((((((.(((.(((((..((((((((..((((((....)).))))....))))))))..)))))..)))(((....)))..........))))))..))).............. ( -49.90) >DroYak_CAF1 50983 120 + 1 UGCAAUGCCGCCUCAUAAACGGUGGCCAUUUGCCUGCCCCAAAUGGCGGCACUUACAAAUGGCACACGUUCGGUGGCCAAGAGGCGGGGUAACUCGGUGGCAUUGCGAAAAGUGUGCAUA .(((((((((((.(((.((((...((((((((..((((((....)).))))....))))))))...))))..)))(((....)))(((....)))))))))))))).............. ( -53.80) >consensus UGCAAAGCCGCCUCAUAAACGGUGGCCAUAUGGCUUCCCCAAACGGCGACACUUACAAAUGGCACACGUUCGGUGGCCAAAAGACGGGGUAAUUCGGUGGCAUUGCGAAAAGUGUGCAAA .((((.((((((...........((((....)))).((((....(....).........((((.(((.....)))))))......))))......)))))).)))).............. (-35.18 = -35.50 + 0.32)

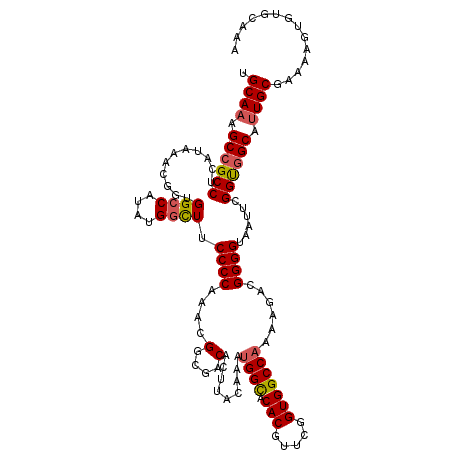
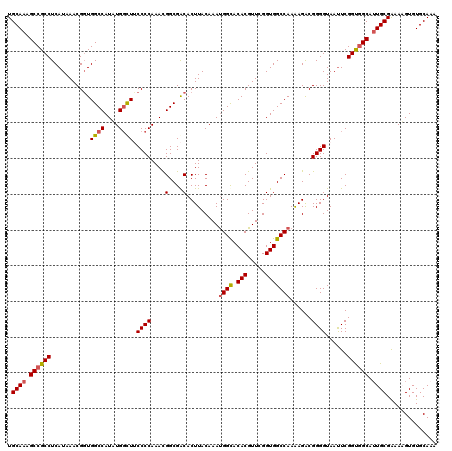
| Location | 10,518,029 – 10,518,149 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -42.62 |
| Consensus MFE | -33.02 |
| Energy contribution | -33.46 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10518029 120 - 22224390 UUUGCACACUUUUCGCAAUGCAACCGAAUUACCCCGUCUUUUGGCCACCGAACGUGUGCCGUUUGUAAGUGUCGCCGUUUGGGGAAGCCAUAUGGCCACCGUUUAUGAGGCGGCUUUGCA ...((((((..((((...((...(((((..((...))..))))).)).)))).))))))....((((((.((((((....(.((..(((....)))..)).)......)))))))))))) ( -35.20) >DroSec_CAF1 49564 120 - 1 UUUGCACACAUUUCGCAAUGCCACCGAAUUACCCCGUCUUUUGGCCACCGAACGUGUACCAUUUGUAAGUGUCGCCGUUUGGGGAAACCAUAUGGCCACCGUUUAUGAGGCGGCUUUGCA ..............((((.(((.((..((.....((.....((((((.((((((.((..((((....))))..))))))))((....))...)))))).))...))..)).))).)))). ( -37.60) >DroSim_CAF1 53088 120 - 1 UUUGCACACAUUUCGCAAUGCCACCGAAUUACCCCGUCUUUUGGCCACCGAACGUGUGCCAUUUGUAAGUGUCGCCGUUUGGGGAAGCCAUAUGGCCACCGUUUAUGAGGCGGCUUUGCA ..............((((.(((..((((..((...))..))))(((.(((((((.(((.((((....)))).))))))))))((..(((....)))..))........)))))).)))). ( -39.10) >DroEre_CAF1 51510 120 - 1 UUUGCACACUUUCCGCACUGCCGCCGAAUUACCCCGCCUUUUGGCCACCGAACGUGUGCCAUUUGUAAGUGCCGCCAUUUGGGGCAGCCAAAUGGCCAACGUUUAUGAGGCGGCUUUGCA ..............(((..((((((..........(((....)))....((((((..((((((((....((((.((....))))))..))))))))..))))))....))))))..))). ( -49.30) >DroYak_CAF1 50983 120 - 1 UAUGCACACUUUUCGCAAUGCCACCGAGUUACCCCGCCUCUUGGCCACCGAACGUGUGCCAUUUGUAAGUGCCGCCAUUUGGGGCAGGCAAAUGGCCACCGUUUAUGAGGCGGCAUUGCA ..............((((((((.((..........(((....)))....(((((((.(((((((((...((((.((....)))))).))))))))))).)))))....)).)))))))). ( -51.90) >consensus UUUGCACACUUUUCGCAAUGCCACCGAAUUACCCCGUCUUUUGGCCACCGAACGUGUGCCAUUUGUAAGUGUCGCCGUUUGGGGAAGCCAUAUGGCCACCGUUUAUGAGGCGGCUUUGCA ..............((((.(((...(........)(((((.(((((((((((((.(((.((((....)))).))))))))))(.....)...))))))........)))))))).)))). (-33.02 = -33.46 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:49 2006