| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,517,306 – 10,517,415 |
| Length | 109 |
| Max. P | 0.690724 |

| Location | 10,517,306 – 10,517,415 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.35 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -26.48 |
| Energy contribution | -27.72 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10517306 109 + 22224390 ---GGGUGUGGCAA-------GUGGGCGGUGGUGCAACACGACUAAGUGCUUUGGCUUGCCGAAUGUAGGCACUUGAACUAUUUUUGUUCUCUCGCC-CACUUACAUACGCGUCGCCAGU ---(.(((((..((-------((((((((((......)))....(((((((((((....)))).....)))))))((((.......))))...))))-))))).))))).)......... ( -41.60) >DroSec_CAF1 48862 109 + 1 ---GGGUGUGGCGU-------GUGGGCGGUGGUGCAACACGACUAAGUGCUUUGGCUUGCCGAAUGUAGGCACUUGAACUAUUUUUGUUCCCUCGCC-CACUUACAUACGCGUCGCCAGU ---.((((..((((-------((((((((.((.((((......((((((((((((....)))).....))))))))........)))).)).)))))-)))......))))..))))... ( -42.04) >DroSim_CAF1 52367 109 + 1 ---GGGUGUGGCAU-------GUGGGCGGUGGUGCAACACGACUAAGUGCUUUGGCUUGCCGAAUGUAGGCACUUGAACUAUUUUUGUUCCCUCGCC-CACUUACAUACGCGUCGCCAGU ---(.(((((....-------((((((((.((.((((......((((((((((((....)))).....))))))))........)))).)).)))))-)))...))))).)......... ( -40.34) >DroEre_CAF1 50873 110 + 1 --ACGGUGUGGCAA-------GUGGGCGAUGGUGCAACACGACUAAGUGCUCUGGCUUGCCGAAUGUAGGCACUUGAACUAUUUUUGUUCUCUCGCC-CACUUACAUACGCAUCGCCAGU --.(((((((..((-------((((((((.(((((..(((......)))...(((....))).......))))).((((.......))))..)))))-))))).....)))))))..... ( -40.00) >DroYak_CAF1 50402 119 + 1 AAAGGGGGUGGCAAUGGGGCUGUGGGCGGUGGUGCAACACGACUAAGUGCUUUGGCUUGCCGAAUGUAGGCACUUGAACUAUUUUUGUUCUCACGCC-CACUUACAUACGCGUCGCCAGU ......((((((.....(...((((((((((......)))((..(((((((((((....)))).....)))))))((((.......)))))).))))-)))...)......))))))... ( -41.30) >DroAna_CAF1 54190 95 + 1 ---UGGUGUA-UAU-------GUGUGUGUGUGUG--------UUAAGUGCUUUGGCAUGCCAAAAGUAGGCACUUGAACUAUUUCUAUGUUCUCGCCGCACUCUCACUCG------CAGU ---.......-..(-------(((.(((.(((((--------..(((((((((((....)))).....)))))))((((.........))))....)))))...))).))------)).. ( -28.00) >consensus ___GGGUGUGGCAA_______GUGGGCGGUGGUGCAACACGACUAAGUGCUUUGGCUUGCCGAAUGUAGGCACUUGAACUAUUUUUGUUCUCUCGCC_CACUUACAUACGCGUCGCCAGU ....((((..((.........((((((((.((.((((......((((((((((((....)))).....))))))))........)))).)).))))).)))........))..))))... (-26.48 = -27.72 + 1.24)

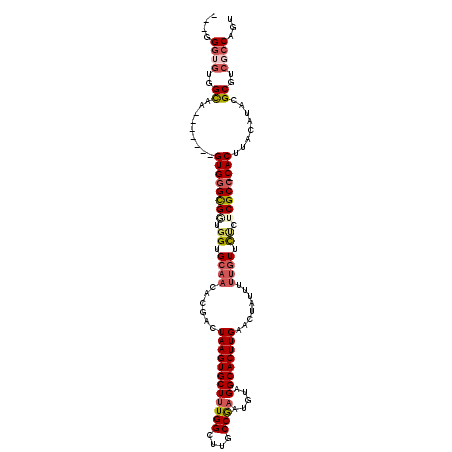

| Location | 10,517,306 – 10,517,415 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.35 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -24.63 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10517306 109 - 22224390 ACUGGCGACGCGUAUGUAAGUG-GGCGAGAGAACAAAAAUAGUUCAAGUGCCUACAUUCGGCAAGCCAAAGCACUUAGUCGUGUUGCACCACCGCCCAC-------UUGCCACACCC--- ..((((.(((....)))(((((-((((...((((.......))))..((((........((....))..(((((......)))))))))...)))))))-------)))))).....--- ( -35.50) >DroSec_CAF1 48862 109 - 1 ACUGGCGACGCGUAUGUAAGUG-GGCGAGGGAACAAAAAUAGUUCAAGUGCCUACAUUCGGCAAGCCAAAGCACUUAGUCGUGUUGCACCACCGCCCAC-------ACGCCACACCC--- ..((((((((....)))..(((-((((..(((((.......))))..((((........((....))..(((((......))))))))))..)))))))-------.))))).....--- ( -37.30) >DroSim_CAF1 52367 109 - 1 ACUGGCGACGCGUAUGUAAGUG-GGCGAGGGAACAAAAAUAGUUCAAGUGCCUACAUUCGGCAAGCCAAAGCACUUAGUCGUGUUGCACCACCGCCCAC-------AUGCCACACCC--- ..((((((((....)))..(((-((((..(((((.......))))..((((........((....))..(((((......))))))))))..)))))))-------.))))).....--- ( -35.30) >DroEre_CAF1 50873 110 - 1 ACUGGCGAUGCGUAUGUAAGUG-GGCGAGAGAACAAAAAUAGUUCAAGUGCCUACAUUCGGCAAGCCAGAGCACUUAGUCGUGUUGCACCAUCGCCCAC-------UUGCCACACCGU-- ....(((.((.(...(((((((-(((((..((((.......))))..((((........((....))..(((((......)))))))))..))))))))-------))))).)).)))-- ( -38.60) >DroYak_CAF1 50402 119 - 1 ACUGGCGACGCGUAUGUAAGUG-GGCGUGAGAACAAAAAUAGUUCAAGUGCCUACAUUCGGCAAGCCAAAGCACUUAGUCGUGUUGCACCACCGCCCACAGCCCCAUUGCCACCCCCUUU ..((((((.((....))..(((-((((((.((((.......))))..((((........((....))..(((((......))))))))))).))))))).......))))))........ ( -35.60) >DroAna_CAF1 54190 95 - 1 ACUG------CGAGUGAGAGUGCGGCGAGAACAUAGAAAUAGUUCAAGUGCCUACUUUUGGCAUGCCAAAGCACUUAA--------CACACACACACAC-------AUA-UACACCA--- ..((------.(.(((.(((((((((..((((.((....))))))..(((((.......))))))))...))))))..--------))).).)).....-------...-.......--- ( -26.30) >consensus ACUGGCGACGCGUAUGUAAGUG_GGCGAGAGAACAAAAAUAGUUCAAGUGCCUACAUUCGGCAAGCCAAAGCACUUAGUCGUGUUGCACCACCGCCCAC_______AUGCCACACCC___ ....((((((((....((((((.(((....((((.......))))...((((.......)))).)))....))))))..))))))))................................. (-24.63 = -25.45 + 0.82)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:45 2006