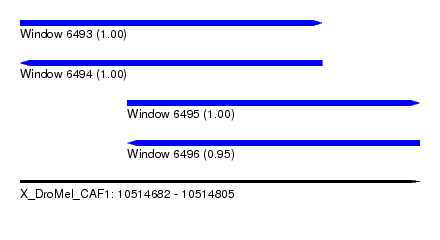
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,514,682 – 10,514,805 |
| Length | 123 |
| Max. P | 0.999968 |

| Location | 10,514,682 – 10,514,775 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.88 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -31.84 |
| Energy contribution | -31.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10514682 93 + 22224390 GAAAGGCUAUUGAAAGAAGAGUCCUUGGAAGGAGAGGGUGGGGUCCUCCAUGCUGGAGGAUGCCGGAUUGGCCAACACUCGGCCAGAUAUCUC ..((((((.((....))..)).)))).....((((...(((.(((((((.....))))))).)))..((((((.......))))))...)))) ( -33.00) >DroSec_CAF1 45983 93 + 1 GAAAGGCUAUUGAAAGAAGAGUCCUUGGAAGGAGAGGGAGGGGUCCUCCAUGCUGGAGGACGCCAGAUUGGCCAACACUCGGCCAGAUAUCUC ..((((((.((....))..)).)))).....((((....((.(((((((.....))))))).))...((((((.......))))))...)))) ( -34.50) >DroSim_CAF1 49320 93 + 1 GAAAGGCUAUUGAAAGAAGAGUCCUUGGAAGGAGAGGGUGGGGUCCUCCAUGCUGGAGGACGCCGGAUUGGCCAACACUCGGCCAGAUAUCUC ..((((((.((....))..)).)))).....((((...(((.(((((((.....))))))).)))..((((((.......))))))...)))) ( -34.50) >DroEre_CAF1 46662 93 + 1 GAAAGGCUAUUGAAAGAAGAGUCCUUGGAAGGAGAGGGUGGGGUCCUCCAUGCUGGAGGACGACGGUUUGGCCAACACUUGGCCAGAUAUCUC ..((((((.((....))..)).)))).....((((...((..(((((((.....)))))))..))(((((((((.....))))))))).)))) ( -34.30) >DroYak_CAF1 47912 93 + 1 GAAAGGCUAUUGAAAGAAGAGUCCUUGGAAGGAGAGGGCGGGGUCCUCCAUGCUGGAGGACGCCGGAUUGGCCAACACUUGGCCAGAUAUCUC ..((((((.((....))..)).)))).....((((...(((.(((((((.....))))))).)))..(((((((.....)))))))...)))) ( -37.70) >consensus GAAAGGCUAUUGAAAGAAGAGUCCUUGGAAGGAGAGGGUGGGGUCCUCCAUGCUGGAGGACGCCGGAUUGGCCAACACUCGGCCAGAUAUCUC ..((((((.((.....)).)).)))).....((((....((.(((((((.....))))))).))...((((((.......))))))...)))) (-31.84 = -31.88 + 0.04)


| Location | 10,514,682 – 10,514,775 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.88 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -29.26 |
| Energy contribution | -29.42 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.00 |
| SVM RNA-class probability | 0.999968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10514682 93 - 22224390 GAGAUAUCUGGCCGAGUGUUGGCCAAUCCGGCAUCCUCCAGCAUGGAGGACCCCACCCUCUCCUUCCAAGGACUCUUCUUUCAAUAGCCUUUC (((.....(((((((...)))))))....((..((((((.....))))))..))...)))((((....))))..................... ( -27.50) >DroSec_CAF1 45983 93 - 1 GAGAUAUCUGGCCGAGUGUUGGCCAAUCUGGCGUCCUCCAGCAUGGAGGACCCCUCCCUCUCCUUCCAAGGACUCUUCUUUCAAUAGCCUUUC .((((...(((((((...)))))))))))((((((((((.....))))))).........((((....))))..............))).... ( -31.40) >DroSim_CAF1 49320 93 - 1 GAGAUAUCUGGCCGAGUGUUGGCCAAUCCGGCGUCCUCCAGCAUGGAGGACCCCACCCUCUCCUUCCAAGGACUCUUCUUUCAAUAGCCUUUC (((.....(((((((...)))))))....((.(((((((.....))))))).))...)))((((....))))..................... ( -30.80) >DroEre_CAF1 46662 93 - 1 GAGAUAUCUGGCCAAGUGUUGGCCAAACCGUCGUCCUCCAGCAUGGAGGACCCCACCCUCUCCUUCCAAGGACUCUUCUUUCAAUAGCCUUUC (((.....((((((.....))))))....(..(((((((.....)))))))..)...)))((((....))))..................... ( -28.00) >DroYak_CAF1 47912 93 - 1 GAGAUAUCUGGCCAAGUGUUGGCCAAUCCGGCGUCCUCCAGCAUGGAGGACCCCGCCCUCUCCUUCCAAGGACUCUUCUUUCAAUAGCCUUUC (((.....((((((.....))))))...(((.(((((((.....))))))).)))..)))((((....))))..................... ( -32.20) >consensus GAGAUAUCUGGCCGAGUGUUGGCCAAUCCGGCGUCCUCCAGCAUGGAGGACCCCACCCUCUCCUUCCAAGGACUCUUCUUUCAAUAGCCUUUC (((.....(((((((...)))))))....((.(((((((.....))))))).))...)))((((....))))..................... (-29.26 = -29.42 + 0.16)


| Location | 10,514,715 – 10,514,805 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -34.34 |
| Energy contribution | -34.58 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10514715 90 + 22224390 GAGGGUGGGGUCCUCCAUGCUGGAGGAUGCCGGAUUGGCCAACACUCGGCCAGAUAUCUCCAGCAGGCUGCACUGCAUCCAAGCAUCAGU------ ..((((((.(((((((.....))))))).))(((((((((.......)))))).....))).((((......))))))))..........------ ( -38.20) >DroSec_CAF1 46016 90 + 1 GAGGGAGGGGUCCUCCAUGCUGGAGGACGCCAGAUUGGCCAACACUCGGCCAGAUAUCUCCAGCAGGCUGCACUGCAUCGAAGCGUCAUU------ ((.(((((((((((((.....))))))).))...((((((.......))))))....)))).((((......)))).))...........------ ( -38.20) >DroSim_CAF1 49353 90 + 1 GAGGGUGGGGUCCUCCAUGCUGGAGGACGCCGGAUUGGCCAACACUCGGCCAGAUAUCUCCAGCAGGCUGCACUGCAUCGGAGCAUCAUU------ .....(((.(((((((.....))))))).)))...(((((.......)))))(((..((((.((((......))))...)))).)))...------ ( -39.10) >DroEre_CAF1 46695 96 + 1 GAGGGUGGGGUCCUCCAUGCUGGAGGACGACGGUUUGGCCAACACUUGGCCAGAUAUCUCCAGCAGGCUGCACUGCAUCCGAGCAUCAGUGUGGAA ((((.((..(((((((.....)))))))..))(((((((((.....))))))))).))))..((((......)))).((((.((....)).)))). ( -41.20) >DroYak_CAF1 47945 96 + 1 GAGGGCGGGGUCCUCCAUGCUGGAGGACGCCGGAUUGGCCAACACUUGGCCAGAUAUCUCCAGCACGCUGCACUGCAUCCGAGCACCAGUGUGGCA ((((.(((.(((((((.....))))))).)))..(((((((.....)))))))...))))......((..(((((...........)))))..)). ( -46.10) >consensus GAGGGUGGGGUCCUCCAUGCUGGAGGACGCCGGAUUGGCCAACACUCGGCCAGAUAUCUCCAGCAGGCUGCACUGCAUCCGAGCAUCAGU______ ((((..((.(((((((.....))))))).))...((((((.......))))))...))))..((((......)))).................... (-34.34 = -34.58 + 0.24)


| Location | 10,514,715 – 10,514,805 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -32.08 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10514715 90 - 22224390 ------ACUGAUGCUUGGAUGCAGUGCAGCCUGCUGGAGAUAUCUGGCCGAGUGUUGGCCAAUCCGGCAUCCUCCAGCAUGGAGGACCCCACCCUC ------.(((.(((......)))...)))..(((((((......(((((((...))))))).)))))))((((((.....)))))).......... ( -34.50) >DroSec_CAF1 46016 90 - 1 ------AAUGACGCUUCGAUGCAGUGCAGCCUGCUGGAGAUAUCUGGCCGAGUGUUGGCCAAUCUGGCGUCCUCCAGCAUGGAGGACCCCUCCCUC ------..............((((......)))).((((.....(((((((...)))))))....((.(((((((.....)))))))))))))... ( -34.30) >DroSim_CAF1 49353 90 - 1 ------AAUGAUGCUCCGAUGCAGUGCAGCCUGCUGGAGAUAUCUGGCCGAGUGUUGGCCAAUCCGGCGUCCUCCAGCAUGGAGGACCCCACCCUC ------...(((((((((..((((......))))))))).))))(((((((...)))))))....((.(((((((.....))))))).))...... ( -39.20) >DroEre_CAF1 46695 96 - 1 UUCCACACUGAUGCUCGGAUGCAGUGCAGCCUGCUGGAGAUAUCUGGCCAAGUGUUGGCCAAACCGUCGUCCUCCAGCAUGGAGGACCCCACCCUC .(((((((((.(((......)))...)))..)).))))(((...((((((.....))))))....)))(((((((.....)))))))......... ( -34.10) >DroYak_CAF1 47945 96 - 1 UGCCACACUGGUGCUCGGAUGCAGUGCAGCGUGCUGGAGAUAUCUGGCCAAGUGUUGGCCAAUCCGGCGUCCUCCAGCAUGGAGGACCCCGCCCUC .((((((((.(.((.(((((((((..(...)..)))....))))))))).)))).)))).....(((.(((((((.....))))))).)))..... ( -40.80) >consensus ______ACUGAUGCUCGGAUGCAGUGCAGCCUGCUGGAGAUAUCUGGCCGAGUGUUGGCCAAUCCGGCGUCCUCCAGCAUGGAGGACCCCACCCUC .........(((((((....((((......))))..))).))))(((((((...)))))))....((.(((((((.....))))))).))...... (-32.08 = -32.24 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:41 2006