
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,508,101 – 10,508,316 |
| Length | 215 |
| Max. P | 0.992676 |

| Location | 10,508,101 – 10,508,216 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 91.65 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -31.60 |
| Energy contribution | -33.20 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10508101 115 + 22224390 CCAGCUAUCCGGGAUGCCAGUUGAUGCUACACACAUAGUUGCCAUUUGCAGCCAGCGAUUUCCAGUUGCUGCAUCAUUCAGAUGCGCGUCGCAUCGCAACGCAUCGCAUUAGCAG ...((((...(.(((((.(((....))).........(((((....((((((.(((........))))))))).......((((((...)))))))))))))))).)..)))).. ( -39.90) >DroSec_CAF1 39492 115 + 1 CCAGCUAUCCGGGAUGCCAGUUGAUGCUACACACAUAGUUGCCAUUUGCAGCCAGCGAUUUCCAGUUGCUGCAUCAUUCAGAUGCGCGUCGCAUUGCAACGCAUCGCAUUAGCAG ...((((...(.(((((((((((((((..........(((((.....))))).((((((.....))))))(((((.....))))))))))).))))....))))).)..)))).. ( -38.10) >DroSim_CAF1 42786 115 + 1 CCAGCUAUCCGGGAUGCCAGUUGAUGCUACACACAUAGUUGCCAUUUGCAGCCAGCGAUUUCCAGUUGCUGCAUCAUUCAGAUGCGCGUCGCAUUGCAACGCAUCGCAUUAGCAG ...((((...(.(((((((((((((((..........(((((.....))))).((((((.....))))))(((((.....))))))))))).))))....))))).)..)))).. ( -38.10) >DroEre_CAF1 40285 115 + 1 CCAGCUAUGCUGCAUGCCAGUUGAUGCUGCACACAUAGUUGCCAUUUGCAGGCAGCGAUUUCCAGUUGCUGCAUCAUUCAGAUGCGCCUCGCAACGCAACGCAUCGCAUUAGCAG ...((((((((((.(((.(((....))))))......(((((...((((((((((((((.....))))))(((((.....))))))))).)))).))))))))..))).)))).. ( -44.70) >DroYak_CAF1 41002 107 + 1 CCAGCUCUCUAGCAUGCCACAUG--------CACAUAGUUGCCAUUUGCUGGCAACGAUUUCCAGUUGCUGCAUCAUUCAGAUGCGCGUCGCAUCGCAACGCAUCGCAUUAGCAG ...((......(((((...))))--------).....(((((....(((.((((((........))))))))).......((((((...)))))))))))))...((....)).. ( -33.70) >consensus CCAGCUAUCCGGGAUGCCAGUUGAUGCUACACACAUAGUUGCCAUUUGCAGCCAGCGAUUUCCAGUUGCUGCAUCAUUCAGAUGCGCGUCGCAUCGCAACGCAUCGCAUUAGCAG ...((((...(.(((((.(((....))).........(((((....(((((.((((........))))))))).......((((((...)))))))))))))))).)..)))).. (-31.60 = -33.20 + 1.60)


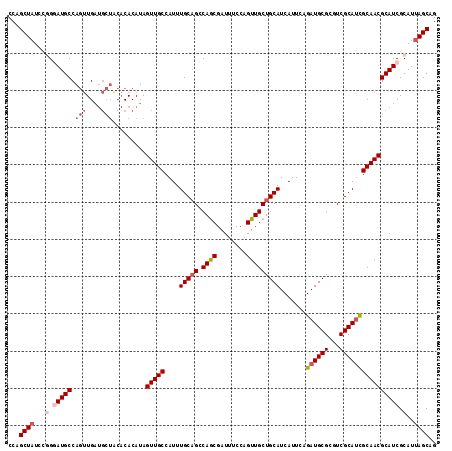
| Location | 10,508,101 – 10,508,216 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.65 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -32.48 |
| Energy contribution | -33.92 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10508101 115 - 22224390 CUGCUAAUGCGAUGCGUUGCGAUGCGACGCGCAUCUGAAUGAUGCAGCAACUGGAAAUCGCUGGCUGCAAAUGGCAACUAUGUGUGUAGCAUCAACUGGCAUCCCGGAUAGCUGG .(((((..((((((((((((...)))))))(((((.....)))))...........)))))..((((((.(((....).))...))))))......)))))..((((....)))) ( -40.90) >DroSec_CAF1 39492 115 - 1 CUGCUAAUGCGAUGCGUUGCAAUGCGACGCGCAUCUGAAUGAUGCAGCAACUGGAAAUCGCUGGCUGCAAAUGGCAACUAUGUGUGUAGCAUCAACUGGCAUCCCGGAUAGCUGG .(((((..((((((((((((...)))))))(((((.....)))))...........)))))..((((((.(((....).))...))))))......)))))..((((....)))) ( -40.90) >DroSim_CAF1 42786 115 - 1 CUGCUAAUGCGAUGCGUUGCAAUGCGACGCGCAUCUGAAUGAUGCAGCAACUGGAAAUCGCUGGCUGCAAAUGGCAACUAUGUGUGUAGCAUCAACUGGCAUCCCGGAUAGCUGG .(((((..((((((((((((...)))))))(((((.....)))))...........)))))..((((((.(((....).))...))))))......)))))..((((....)))) ( -40.90) >DroEre_CAF1 40285 115 - 1 CUGCUAAUGCGAUGCGUUGCGUUGCGAGGCGCAUCUGAAUGAUGCAGCAACUGGAAAUCGCUGCCUGCAAAUGGCAACUAUGUGUGCAGCAUCAACUGGCAUGCAGCAUAGCUGG ..((((.(((.....(((((.((((.(((((((((.....)))))(((...........)))))))))))...)))))..(((((((((......)).))))))))))))))... ( -42.70) >DroYak_CAF1 41002 107 - 1 CUGCUAAUGCGAUGCGUUGCGAUGCGACGCGCAUCUGAAUGAUGCAGCAACUGGAAAUCGUUGCCAGCAAAUGGCAACUAUGUG--------CAUGUGGCAUGCUAGAGAGCUGG .(((((((((((((((((((...)))))))(((((.....)))))...........)))(((((((.....))))))).....)--------))).))))).(((....)))... ( -41.20) >consensus CUGCUAAUGCGAUGCGUUGCGAUGCGACGCGCAUCUGAAUGAUGCAGCAACUGGAAAUCGCUGGCUGCAAAUGGCAACUAUGUGUGUAGCAUCAACUGGCAUCCCGGAUAGCUGG .(((((..((((((((((((...)))))))(((((.....)))))...........)))))..((((((.(((....).))...))))))......)))))..((((....)))) (-32.48 = -33.92 + 1.44)


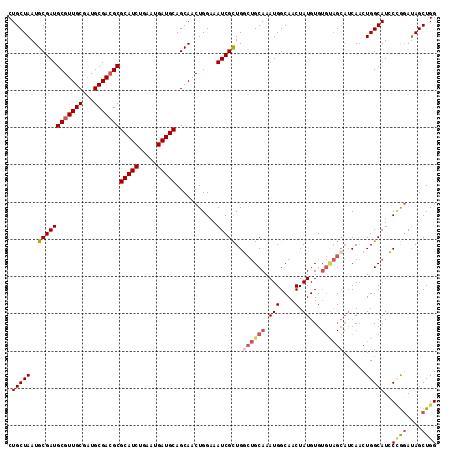
| Location | 10,508,216 – 10,508,316 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.20 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10508216 100 - 22224390 UACCCCCACAGCAAAUCCUCCCGCUCUUGCUGUGGCAACUCCUCCAUUCAUCAG---CUGCCAGUUGUCAAUUGUCAGUUGUCACUUGGCUGCCUCGUGUGGC .....(((((((((............)))))))))........((((......(---(.(((((.((.((((.....)))).)).))))).)).....)))). ( -29.80) >DroSec_CAF1 39607 100 - 1 UCCUCCCACAGCAAAUCCUCCCGCUACUGAUGUGGCAUCUCCUCCAUUCAUCAG---CUGCCAGUUGUCAAUUGUCAGUUGUCACUUGGCUGCCUCGUGUGGC .....(((((............(((((....))))).................(---(.(((((.((.((((.....)))).)).))))).))....))))). ( -27.30) >DroSim_CAF1 42901 100 - 1 UACUCCCACAGCAAAUCCUCCCGCUACUGAUGUGGCAUCUCCUCCAUUCAUCAG---CUGCCAGUUGUCAAUUGUCAGUUGUCACUUGGCUGCCUCGUGUGGC .....(((((............(((((....))))).................(---(.(((((.((.((((.....)))).)).))))).))....))))). ( -27.30) >DroEre_CAF1 40400 100 - 1 ACCUCCCACACCCAAUCCUCCCGCUCAUGCUGUGGCAUCUCCACCAUUCAUCAG---CUGCCAGUUGUCAAUUGUCAGUUGUCACUUGGCUGCCUCGUGUGGC .....((((((...........(((.(((..((((........)))).))).))---).(((((.((.((((.....)))).)).)))))......)))))). ( -28.90) >DroAna_CAF1 43532 94 - 1 ACCGCCCUUGUUCAUGCCACACACUCAUCAUCUGG---------CAGUCGUCAGUCGUUGUCGGUUGUCAAUUGUCAGUUGUCACUUGGCUGCCUCGUGUGGC ...............(((((((.........((((---------((((.(.((..((....))..)).).))))))))..(((....)))......))))))) ( -29.20) >consensus UCCUCCCACAGCAAAUCCUCCCGCUCAUGAUGUGGCAUCUCCUCCAUUCAUCAG___CUGCCAGUUGUCAAUUGUCAGUUGUCACUUGGCUGCCUCGUGUGGC .....(((((..........((((.......))))........................(((((.((.((((.....)))).)).))))).......))))). (-17.80 = -17.60 + -0.20)


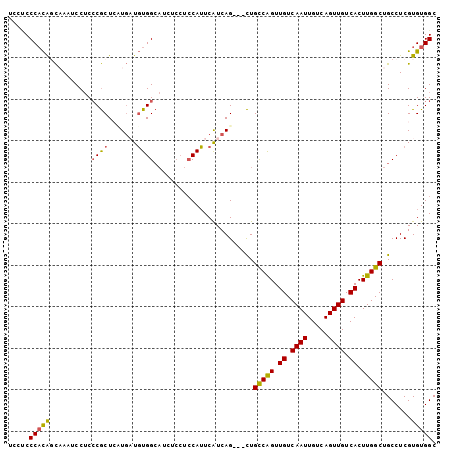
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:30 2006