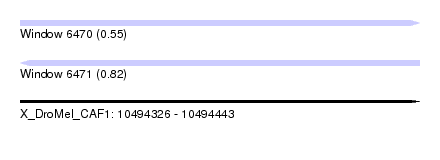
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 10,494,326 – 10,494,443 |
| Length | 117 |
| Max. P | 0.821461 |

| Location | 10,494,326 – 10,494,443 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -23.46 |
| Energy contribution | -24.35 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10494326 117 + 22224390 GUCCUUUUGUCCAACUGCUUGUGGCGCUUUCGCAAUUUGAUUUAUGUGUUGCC---CCCAAAACAUUCAUUAAGAUAUAAUCAUAACGCAUAGAAAGCGCACACAAACAGGAUGAGUAUG .....((..(((...((.(((((((((((((((...(((((..((((.(((..---..))).))))..)))))(((...))).....))...)))))))).))))).)))))..)).... ( -30.20) >DroSec_CAF1 25502 117 + 1 GUCCUUUUGUCCAACAGCUUGUGGCGCUUUCGCAAUUUGAUUUAUGUGUUGCC---CCCAAAACAUUCAUUAAGAUAUAAUCAUAACGCAUAGAAAGCGCACACAAACAGGAUGAGUAUG .....((..(((....(.(((((((((((((((...(((((..((((.(((..---..))).))))..)))))(((...))).....))...)))))))).))))).).)))..)).... ( -28.50) >DroSim_CAF1 26014 117 + 1 GUCCUUUUGUCCAACUGCUUGUGGCGCUUUCGCAAUUUGAUUUAUGUGUUGCC---CCCAAAACAUUCAUUAAGAUAUAAUCAUAACGCAUAGAAAGCGCACACAAACAGGAUGAGUAUG .....((..(((...((.(((((((((((((((...(((((..((((.(((..---..))).))))..)))))(((...))).....))...)))))))).))))).)))))..)).... ( -30.20) >DroEre_CAF1 27125 117 + 1 GUCCUUUUGUCCAACUGCUUGUGGCGCUUUCGCAAUUUGAUUUAUGUGUUGCC---CCCAAAACAUUCAUUAAGAUAUAAUCAUAACGCAUAGAAAGCGCACACAAACAGGAUGAGUAUG .....((..(((...((.(((((((((((((((...(((((..((((.(((..---..))).))))..)))))(((...))).....))...)))))))).))))).)))))..)).... ( -30.20) >DroYak_CAF1 27430 117 + 1 GUCCUUGUGUCCAACUGCUUGUGGCGCUUUCGCAAUUUGAUUUAUGUGUUGCC---CCCAAAACAUUCAUUAAGAUAUAAUCAUAACGCAUAGAAAGCGCACACAAACAGGAUGAGUACG ......(((..((.(((.(((((((((((((((...(((((..((((.(((..---..))).))))..)))))(((...))).....))...)))))))).))))).)))..))..))). ( -32.70) >DroAna_CAF1 28397 114 + 1 GUCCUUUUGUCCAACUGCUUGUGGCGUUUUCGCAAUUUGAUUUAUGUCCUGGCCCCCCAAAAACAUUCAUUAAGAUAUAAUCAUAACGCAUACAAGGCGCAAACACUCACACUC------ .....(((((((...(((((((((((....)))..((((((..((((..(((....)))...))))..)))))).......))))).))).....)).)))))...........------ ( -18.80) >consensus GUCCUUUUGUCCAACUGCUUGUGGCGCUUUCGCAAUUUGAUUUAUGUGUUGCC___CCCAAAACAUUCAUUAAGAUAUAAUCAUAACGCAUAGAAAGCGCACACAAACAGGAUGAGUAUG ..............(((.(((((((((((((((...(((((..((((.(((.......))).))))..)))))(((...))).....))...)))))))).))))).))).......... (-23.46 = -24.35 + 0.89)

| Location | 10,494,326 – 10,494,443 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -27.50 |
| Energy contribution | -28.87 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 10494326 117 - 22224390 CAUACUCAUCCUGUUUGUGUGCGCUUUCUAUGCGUUAUGAUUAUAUCUUAAUGAAUGUUUUGGG---GGCAACACAUAAAUCAAAUUGCGAAAGCGCCACAAGCAGUUGGACAAAAGGAC .....((...(((((((((.((((((((...(((...(((((...((.....))((((.(((..---..))).)))).)))))...))))))))))))))))))))...))......... ( -34.90) >DroSec_CAF1 25502 117 - 1 CAUACUCAUCCUGUUUGUGUGCGCUUUCUAUGCGUUAUGAUUAUAUCUUAAUGAAUGUUUUGGG---GGCAACACAUAAAUCAAAUUGCGAAAGCGCCACAAGCUGUUGGACAAAAGGAC ........(((.(((((((.((((((((...(((...(((((...((.....))((((.(((..---..))).)))).)))))...))))))))))))))))))....)))......... ( -33.20) >DroSim_CAF1 26014 117 - 1 CAUACUCAUCCUGUUUGUGUGCGCUUUCUAUGCGUUAUGAUUAUAUCUUAAUGAAUGUUUUGGG---GGCAACACAUAAAUCAAAUUGCGAAAGCGCCACAAGCAGUUGGACAAAAGGAC .....((...(((((((((.((((((((...(((...(((((...((.....))((((.(((..---..))).)))).)))))...))))))))))))))))))))...))......... ( -34.90) >DroEre_CAF1 27125 117 - 1 CAUACUCAUCCUGUUUGUGUGCGCUUUCUAUGCGUUAUGAUUAUAUCUUAAUGAAUGUUUUGGG---GGCAACACAUAAAUCAAAUUGCGAAAGCGCCACAAGCAGUUGGACAAAAGGAC .....((...(((((((((.((((((((...(((...(((((...((.....))((((.(((..---..))).)))).)))))...))))))))))))))))))))...))......... ( -34.90) >DroYak_CAF1 27430 117 - 1 CGUACUCAUCCUGUUUGUGUGCGCUUUCUAUGCGUUAUGAUUAUAUCUUAAUGAAUGUUUUGGG---GGCAACACAUAAAUCAAAUUGCGAAAGCGCCACAAGCAGUUGGACACAAGGAC .((..(((..(((((((((.((((((((...(((...(((((...((.....))((((.(((..---..))).)))).)))))...)))))))))))))))))))).)))..))...... ( -36.00) >DroAna_CAF1 28397 114 - 1 ------GAGUGUGAGUGUUUGCGCCUUGUAUGCGUUAUGAUUAUAUCUUAAUGAAUGUUUUUGGGGGGCCAGGACAUAAAUCAAAUUGCGAAAACGCCACAAGCAGUUGGACAAAAGGAC ------(..((((.((((((.(((........(((((.(((...))).))))).(((((((.((....)))))))))..........))).))))))))))..)................ ( -25.10) >consensus CAUACUCAUCCUGUUUGUGUGCGCUUUCUAUGCGUUAUGAUUAUAUCUUAAUGAAUGUUUUGGG___GGCAACACAUAAAUCAAAUUGCGAAAGCGCCACAAGCAGUUGGACAAAAGGAC ..........(((((((((.((((((((...(((...(((((...((.....))((((.(((.......))).)))).)))))...)))))))))))))))))))).............. (-27.50 = -28.87 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:18 2006